Visualizing Dynamic Biofilm Processes: How High-Speed AFM is Revolutionizing Microbiology and Drug Development
This article explores the transformative role of High-Speed Atomic Force Microscopy (HS-AFM) in visualizing the dynamic processes of biofilm formation and behavior in real-time.
Visualizing Dynamic Biofilm Processes: How High-Speed AFM is Revolutionizing Microbiology and Drug Development
Abstract
This article explores the transformative role of High-Speed Atomic Force Microscopy (HS-AFM) in visualizing the dynamic processes of biofilm formation and behavior in real-time. Tailored for researchers, scientists, and drug development professionals, it covers the foundational principles of HS-AFM, detailing its capability to capture nanoscale structural and functional changes at sub-second resolution under physiological conditions. The content provides a methodological guide for application, addresses common troubleshooting and optimization challenges, and offers a comparative analysis with other biomechanical characterization techniques. By synthesizing the latest research and technical advances, this article serves as a comprehensive resource for leveraging HS-AFM to develop novel anti-biofilm strategies and therapeutics.
Unveiling the Biofilm Lifecycle: Foundational Principles of High-Speed AFM Imaging
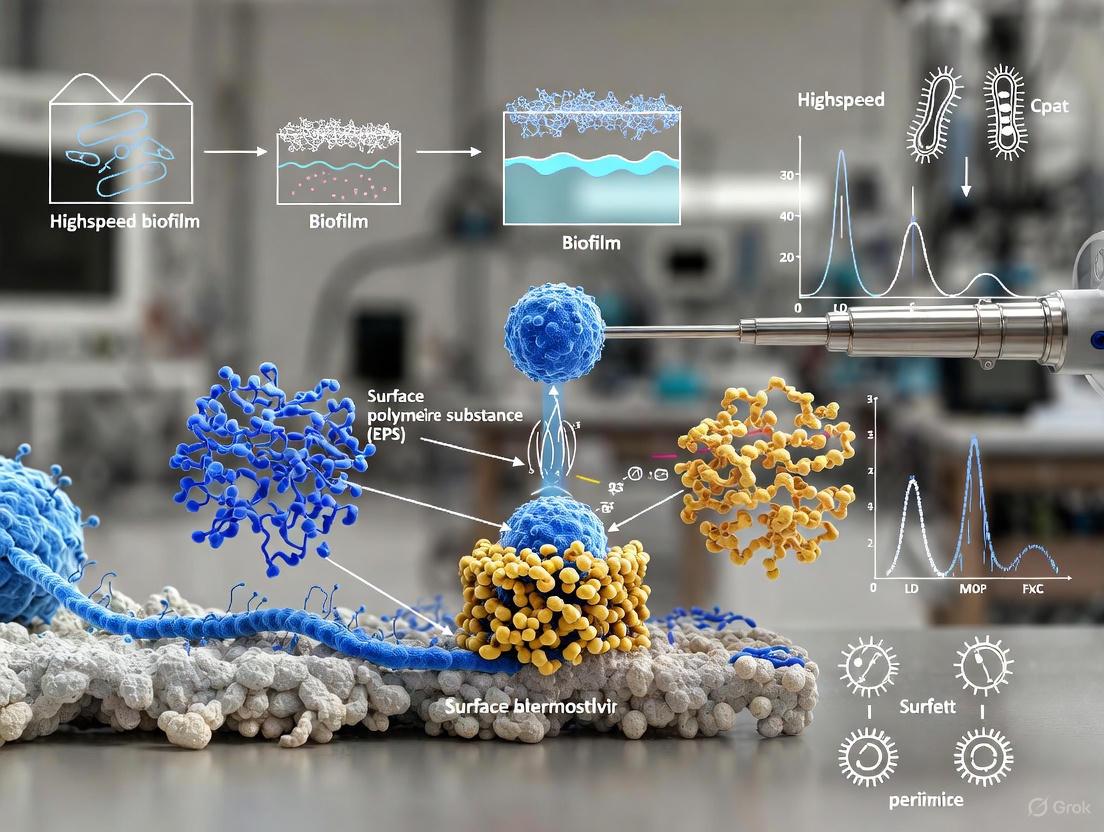
Biofilms are complex, three-dimensional microbial communities encased in a self-produced matrix of extracellular polymeric substances (EPS). Their remarkable resilience and multifaceted functions stem from intricate microbial interactions, including enhanced substrate metabolism, energy conversion, cellular communication, and horizontal gene transfer [1]. However, this complexity also presents a significant research challenge: biofilms are inherently heterogeneous and dynamic, characterized by spatial and temporal variations in structure, composition, density, and metabolic activity [2]. These variations are influenced by microbial species, environmental conditions, surface properties, and microbial interactions, all of which contribute to biofilm stability and resilience [2].
Traditional imaging technologies face fundamental obstacles in capturing this dynamic complexity. Conventional methods, including Atomic Force Microscopy (AFM), Confocal Laser Scanning Microscopy (CLSM), and Scanning Electron Microscopy (SEM), often involve labor-intensive data acquisition, complex analysis workflows, low reproducibility, and limited throughput [1]. More critically, these methods frequently fall short in capturing the intricate spatial and functional dynamics of living biofilms and can introduce artifacts that compromise result fidelity [1]. The core issue is a mismatch in timescales; the rapid, transient processes of biofilm formation occur much faster than the acquisition speed of conventional, high-resolution imaging tools.
Limitations of Conventional Biofilm Imaging Methods
The limitations of traditional imaging techniques become starkly apparent when applied to the study of dynamic biofilm processes. A comparative analysis of their shortcomings is presented in the table below.
Table 1: Limitations of Traditional Imaging Techniques for Dynamic Biofilm Studies
| Imaging Technique | Key Limitations for Dynamic Studies | Impact on Biofilm Data Fidelity |
|---|---|---|
| Atomic Force Microscopy (AFM) | Small imaging area (<100 µm); Slow scanning process; Labor-intensive operation; Limited capture of dynamic structural changes [2]. | Inability to link nanoscale features to macroscale organization; Misses rapid, transient events in biofilm development. |
| Confocal Laser Scanning Microscopy (CLSM) | Labor-intensive data acquisition; Limited throughput; Potential for photodamage during long-term imaging [1]. | Incomplete representation of rapid 3D architectural changes; Altered biofilm physiology due to light exposure. |
| Scanning Electron Microscopy (SEM) | Requires sample dehydration and metallic coating; Complex preparation causes sample distortion and Eps collapse [3]. | Provides only static snapshots of fixed samples; non-physiological conditions preclude live, dynamic monitoring. |
| Raman Spectroscopy (RS) | Fluorescence interference; Low reproducibility; Requires high laser power risking photodamage [1]. | Hinders reliable, repeated measurements of chemical dynamics within living biofilms. |
A primary technical hurdle for conventional AFM is its limited scan range, which restricts the ability to link critical smaller-scale features—such as individual cell morphology and nanomechanical properties—to the emergent functional architecture of the biofilm at the macroscale [2]. Furthermore, the heterogeneity of biofilms, with their intricate networks of pores, channels, and cell clusters often falling below the resolution of conventional technologies, results in an incomplete representation of true biofilm morphology [1]. These limitations collectively underscore that traditional methods are ill-suited for observing biofilm processes as they unfold in real-time.
High-Speed and Automated AFM: A Paradigm Shift for Dynamic Analysis
Recent technological advancements are overcoming these historical barriers. The development of large area automated AFM represents a transformative approach, enabling the capture of high-resolution images over millimeter-scale areas, a significant leap from the traditional sub-100µm range [2]. This innovation directly addresses the scale mismatch, allowing researchers to correlate cellular-level events with community-level organization.
Automation is a critical component of this new paradigm. Machine Learning (ML) and Artificial Intelligence (AI) are now being integrated to automate the scanning process, including sample region selection, scanning optimization, and probe conditioning [2]. This AI-driven automation minimizes user intervention, allowing for continuous, multi-day experiments and overcoming the labor-intensive nature of traditional AFM operation [2]. The application of deep learning algorithms, such as convolutional neural networks (CNNs) for image segmentation and generative adversarial networks for image reconstruction, effectively enhances resolution, reduces artifacts, and captures precise spatiotemporal biofilm architecture previously unattainable [1].
Table 2: Quantitative Capabilities of Advanced AFM Modalities for Biofilm Research
| AFM Modality | Key Quantitative Outputs | Application in Dynamic Monitoring |
|---|---|---|
| High-Speed/Large-Area AFM | Cell count, confluency, cell shape/orientation over mm² areas [2]. | Tracking early bacterial adhesion patterns and cluster formation in real-time. |
| Force Spectroscopy | Adhesion forces (nN), elastic moduli (Young's modulus), turgor pressure [4]. | Mapping spatiotemporal variations in biofilm mechanical properties during treatment. |
| Tapping Mode in Liquid | Topographical height (nm), surface roughness (Rq), phase imaging for material contrast [4]. | In-situ, non-destructive 3D visualization of hydrated biofilm matrix development. |
These advanced AFM modalities facilitate a multiparametric investigation. Force spectroscopy allows for the measurement of interaction forces between biomolecules and nanomechanical properties like stiffness, which influence antimicrobial penetration and biofilm removal [3]. When combined with the ability to operate in liquid under physiological conditions, these new AFM platforms enable a non-invasive, quantitative, and comprehensive analysis of living biofilms, from initial attachment to mature community responses to environmental stresses.
Essential Research Reagent Solutions for Biofilm AFM
Successful high-speed AFM imaging of dynamic biofilms relies on a suite of specialized reagents and materials for sample preparation and analysis.
Table 3: Research Reagent Solutions for High-Speed AFM Biofilm Studies
| Research Reagent / Material | Function in Experimental Protocol |
|---|---|
| PFOTS-treated Glass | Creates a hydrophobic surface to promote and study controlled bacterial adhesion for early attachment studies [2]. |
| Polydimethylsiloxane (PDMS) Stamps | Microfabricated stamps with specific pit dimensions (1.5–6 µm) for secure, organized mechanical immobilization of microbial cells [4]. |
| Poly-L-Lysine | A chemical immobilization agent that promotes electrostatic binding of cells to substrates like mica or glass [4]. |
| Osmium Tetroxide (OsOâ‚„) | A staining and fixation agent used in sample preparation protocols to preserve lipid structures and enhance SEM/AFM image quality [3]. |
| Ruthenium Red (RR) & Tannic Acid (TA) | Used in customized staining protocols to specifically stabilize and visualize the extracellular polymeric substance (EPS) matrix [3]. |
| Ionic Liquid (IL) Treatment | Applied to samples to enhance electrical conductivity, reducing charging artifacts in SEM and enabling higher-resolution imaging without metal coating [3]. |
| Pantoea sp. YR343 | A gram-negative, motile model bacterium with peritrichous flagella, ideal for studying the role of appendages in early biofilm assembly [2]. |
Experimental Protocol: High-Speed AFM for Early Biofilm Assembly
Objective: To visualize and quantify the initial attachment and organization of bacterial cells on a surface using automated large-area AFM.
Materials:
- Bacterial strain (e.g., Pantoea sp. YR343) [2].
- Growth medium appropriate for the selected strain.
- PFOTS-treated glass coverslips or PDMS immobilization stamps [2] [4].
- Atomic Force Microscope with large-area scanning capability and an appropriate liquid cell.
- Fluidic system for inoculation and rinsing.
Procedure:
- Surface Preparation: Prepare PFOTS-treated glass coverslips to create a standardized hydrophobic surface for bacterial attachment [2].
- Sample Inoculation: Place the prepared coverslip in a Petri dish and inoculate with bacterial cells suspended in a liquid growth medium.
- Controlled Incubation: Incubate the sample for a selected, short time period (e.g., ~30 minutes) to allow for initial cell attachment [2].
- Gentle Rinsing: Carefully remove the coverslip from the Petri dish and gently rinse with a buffer solution to remove non-adherent planktonic cells.
- AFM Mounting: Securely mount the rinsed sample in the AFM liquid cell, ensuring it is fully hydrated in buffer to maintain physiological conditions.
- Automated Large-Area Scanning:
- Program the AFM to automatically acquire multiple high-resolution images across a millimeter-scale area of interest.
- Utilize machine learning algorithms for optimal site selection and scanning parameter adjustment [2].
- Image Stitching and Analysis:
- Apply computational stitching algorithms to reconstruct a seamless, high-resolution map of the entire scanned area.
- Use machine learning-based image segmentation to automatically detect cells, classify features, and extract quantitative parameters (e.g., cell count, confluency, cell orientation, flagellar presence) [2].
Diagram 1: High-Speed AFM Workflow for Early Biofilm Assembly
The critical need for speed in biofilm imaging is unequivocally addressed by the advent of high-speed and automated AFM technologies. By overcoming the fundamental limitations of traditional imaging—specifically small scan areas, slow acquisition rates, and labor-intensive operation—these advanced methods unlock the potential to observe and quantify the dynamic processes of biofilm formation and adaptation in real-time. The integration of artificial intelligence and machine learning is pivotal, transforming AFM from a manual, single-image tool into an automated, quantitative platform capable of millimeter-scale analysis.
The future of dynamic biofilm research lies in the continued integration of these high-speed imaging capabilities with other multimodal data streams. Combining the nanoscale topographical and mechanical data from AFM with the chemical information from techniques like Raman spectroscopy, all underpinned by AI-driven analysis, promises a holistic and systems-level understanding of biofilm biology. This powerful synergy will ultimately accelerate the development of targeted interventions to control problematic biofilms across medical, industrial, and environmental contexts.
High-speed atomic force microscopy (HS-AFM) has emerged as a transformative tool for directly observing dynamic biological processes at the nanoscale, including the assembly and remodeling of biofilms, in near-physiological conditions. Unlike conventional AFM, HS-AFM captures data at sub-second to second temporal resolution, enabling researchers to visualize molecular and cellular dynamics in real-time. This capability is particularly valuable for studying biofilm development, where structural changes occur over timescales ranging from milliseconds to hours. The performance of an HS-AFM system hinges on three critical components: miniaturized cantilevers that enable fast response times, high-speed scanners that facilitate rapid tip positioning, and sensitive detectors that accurately measure cantilever deflection. This application note details the core principles, specifications, and operational protocols for these components, with a specific focus on their application in dynamic biofilm research for drug development and antimicrobial strategy evaluation.
Core Component 1: Cantilevers
The cantilever is the primary force sensor in AFM and the most critical component for achieving high-speed imaging. Its physical properties directly determine the imaging speed, sensitivity, and signal-to-noise ratio (SNR).
Traditional vs. Advanced Cantilever Designs
Conventional HS-AFM employs miniaturized beam-shaped cantilevers to achieve high resonant frequencies and low spring constants. However, further miniaturization of these beams faces a fundamental trade-off: as the cantilever becomes smaller to increase speed, the area available for laser reflection decreases, leading to a lower SNR [5].
A recent breakthrough, the seesaw cantilever, presents a paradigm shift in design. This architecture decouples the mechanical element (torsional hinges) from the laser-reflective element (a rigid board) [5]. The key advantages of this design are:
- Enhanced SNR: The reflective board can be optimized for laser reflection independent of the mechanical properties, leading to a superior signal-to-noise ratio [5].
- Independent Parameter Tuning: The stiffness is precisely tuned via the hinge dimensions (length, width, thickness), while the board size is optimized for optical detection and hydrodynamic drag [5].
- High Angular Sensitivity: The shortened distance between the tip and the hinges enhances sensitivity to tip-sample interactions [5].
Table 1: Comparison of Traditional and Seesaw Cantilever Designs for HS-AFM
| Feature | Traditional Beam Cantilever | Seesaw Cantilever |
|---|---|---|
| Design Principle | Bending beam | Swinging board on torsional hinges |
| Key Limitation | Laser reflection area decreases with miniaturization, limiting SNR | More complex fabrication process |
| Laser Reflectivity | Coupled to beam dimensions; compromised in miniaturized versions | Decoupled; board can be optimized for high reflectivity |
| Stiffness Tuning | Adjusted via length, width, and thickness of the entire beam | Precisely tuned via the dimensions of the torsional hinges |
| Reported Performance | Soft and fast, but SNR is a limiting factor at extreme miniaturization | Surpasses best beam cantilevers in sensitivity; matches imaging performance [5] |
Cantilever Selection and Fabrication Protocol
Objective: To select and/or fabricate a high-speed cantilever suitable for imaging dynamic biofilm processes with high temporal and spatial resolution.
Materials:
- Cantilever Chips: Silicon or silicon nitride cantilevers. For seesaw prototypes, a larger silicon nitride cantilever body (e.g., 200 µm long, 30 µm wide, 0.4 µm thick) coated with 5 nm Ti and 40 nm Au can be used as a platform for fabrication [5].
- Fabrication System: Focused Ion Beam (FIB) milling system for prototyping seesaw cantilevers [5].
- Tip Growth System: Electron Beam Deposition (EBD) for growing high-aspect-ratio tips (~2.5 µm long) [5].
Procedure:
- Design Specification:
- Define target resonant frequency (typically > 1 MHz in liquid) and spring constant (typically 0.1-0.2 N/m for biological samples) based on imaging requirements.
- For seesaw cantilevers, determine the board dimensions (e.g., 5 µm x 10 µm or 5 µm x 5 µm) and hinge geometry (e.g., 1 µm long x 0.2 µm wide) to achieve desired mechanical properties [5].
FIB Milling Fabrication (for Seesaw Cantilevers) [5]:
- a. Initial Trench Milling: Use FIB to mill an inverted Î -shaped trench near the cantilever's supporting base.
- b. Second Trench Milling: Mill a second Π-shaped trench opposite the first, leaving a 1 µm x 1 µm gap that will form the torsional hinges.
- c. Material Removal: Mill lateral trenches toward the periphery of the silicon nitride plate, leaving a small bar for structural integrity.
- d. Final Release: Mill the remaining peripheral regions to release the top part, creating the free-standing seesaw cantilever.
Tip Integration:
- Using EBD, grow a sharp, high-aspect-ratio tip (~2.5 µm in length) at the distal periphery of the cantilever board. The tip should be placed centrally along the board's long axis [5].
Validation:
- Perform finite element analysis to simulate the oscillation behavior and resonant frequency.
- Characterize the final cantilever using scanning electron microscopy (SEM) to verify dimensions and tip placement.
- Experimentally determine the resonant frequency and stiffness in the intended fluid environment.
Core Component 2: Scanners
High-speed scanners position the sample or the probe with nanometer precision at high velocities. Their performance is critical for minimizing image distortion and achieving high frame rates.
Scanner Technology and Nonlinear Compensation
Piezoelectric actuators (PEAs) are the standard for AFM scanners due to their high precision, fast response, and large actuation force [6]. However, their intrinsic nonlinear behaviors, primarily hysteresis and creep, pose significant challenges for accurate scanning, especially at high speeds. Hysteresis causes the scanner's displacement to depend on its previous motion, leading to distorted images [6].
Advanced control strategies are essential to mitigate these effects. While feedback control with position sensors is effective, it can be limited by sensor noise and actuator bandwidth. A promising alternative is model-based feedforward compensation, which uses a mathematical model to predict and cancel out the nonlinearities without requiring physical sensors [6].
Recent research demonstrates the efficacy of using a variant DenseNet-type neural network to model and compensate for PEA hysteresis. This approach uses skip connections to improve information flow, preventing the "vanishing gradient" problem in deep networks and achieving a remarkably low relative root-mean-square (RMS) error of less than 0.1% [6]. This method simplifies the modeling by using separate models for the scanner's forward and backward movements.
Table 2: Key Characteristics of a High-Speed Piezoelectric Scanner
| Parameter | Specification | Importance for HS-AFM |
|---|---|---|
| Scan Range (XY) | ~27 × 27 µm² | Determines the maximum imaging area per frame [6] |
| Resonant Frequency | As high as possible (e.g., >10 kHz) | Limits the maximum achievable scan speed |
| Hysteresis | Compensated to <0.1% RMS error | Critical for accurate spatial positioning and image fidelity [6] |
| Actuator Material | Lead Zirconate Titanate (PZT) Ceramic | Provides the piezoelectric effect for precise motion [6] |
| Control Strategy | DenseNet-type Neural Network Feedforward | Compensates for nonlinearities without position sensors, enabling high-speed operation [6] |
Protocol: Neural Network-Based Scanner Modeling
Objective: To create an accurate inverse hysteresis model of a piezoelectric scanner for high-precision, high-speed positioning.
Materials:
- Piezoelectric Scanner: A custom or commercial HS-AFM scanner with piezoelectric actuators (e.g., Thorlabs PK4FQP2 PEA) [6].
- High-Voltage Amplifier: Small signal bandwidth of >100 kHz.
- Displacement Sensor: Capacitive sensor (e.g., MicroSense 5504) with gauging module for measuring actual scanner displacement [6].
- Data Acquisition System: FPGA module (e.g., National Instruments PXI-7853R) for input/output control and data acquisition [6].
Procedure:
- Data Collection:
- Drive each axis of the XY scanner with triangular input signals of varying amplitudes (e.g., 25 V, 50 V, 75 V, 100 V).
- For each input trajectory, record the corresponding output displacement using the capacitive sensor. Collect a minimum of 1000 samples each for the forward and backward directions.
- Repeat the entire procedure multiple times (e.g., 20x) to reduce measurement error and ensure a robust dataset [6].
Model Construction:
- Implement a variant DenseNet-type neural network. Utilize skip connections to link non-adjacent layers, ensuring robust gradient flow during training.
- Construct two separate models: one dedicated to mapping the forward direction data and another for the backward direction.
Training and Validation:
- Split the collected data into training and testing sets (e.g., 80/20 split).
- Train the models to predict the input voltage required to achieve a desired displacement, effectively learning the inverse hysteresis.
- Validate model performance by comparing the scanner's actual displacement to the desired trajectory when the model is used for feedforward control. The target is to reduce the relative RMS error to below 0.1% [6].
Core Component 3: Detectors
The deflection detection system converts the minute motion of the cantilever into a measurable electrical signal. A high-bandwidth, low-noise detector is essential for resolving high-speed cantilever oscillations.
Optical Beam Deflection System
The optical beam deflection (OBD) system is the most widely used detection method. Its core components are a laser diode, a focusing lens, and a position-sensitive photodetector (PSPD) [7]. The principle of operation is as follows:
- A laser beam is focused onto the back of the cantilever.
- The reflected beam is directed onto a PSPD.
- Cantilever bending (deflection) or swinging (torsion) causes the position of the reflected laser spot on the PSPD to shift.
- The PSPD converts this positional change into a voltage signal proportional to the cantilever's motion [7].
The seesaw cantilever's design, with its large, rigid reflective board, provides a significant advantage in this system by offering a stable and optimized surface for laser reflection, thereby increasing the signal-to-noise ratio compared to miniaturized beam cantilevers [5].
Workflow: Integrated HS-AFM Imaging of Biofilm Dynamics
The following diagram illustrates the synergistic interaction of the three core components during a typical HS-AFM experiment for biofilm imaging.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for HS-AFM Biofilm Research
| Item | Function/Application | Examples & Notes |
|---|---|---|
| Silicon Nitride Cantilevers | Base material for fabricating durable, high-frequency cantilevers. | Bruker AFM probes; used as a platform for FIB milling of custom seesaw cantilevers [5] [7]. |
| Focused Ion Beam (FIB) System | Precision milling and prototyping of advanced cantilever designs (e.g., seesaw). | Critical for R&D of next-generation cantilevers with decoupled reflective/mechanical functions [5]. |
| High-Speed Piezoelectric Scanner | Provides rapid, precise sample or probe positioning for fast imaging. | XY scanners with resonant frequencies >10 kHz; often require neural network control for hysteresis compensation [6]. |
| Supported Lipid Bilayers (SLBs) | Mimic bacterial or host cell membranes for studying membrane protein dynamics within biofilms. | Composed of DOPC or E. coli lipids; protein mobility varies with lipid composition [8]. |
| Pantoea sp. YR343 | A model gram-negative bacterium for studying early-stage biofilm assembly and structure. | Rod-shaped, motile bacterium; forms honeycomb-like patterns observable via HS-AFM [9]. |
| FluidFM Technology | Enables biofilm-scale force spectroscopy by immobilizing biofilm-coated beads on a cantilever. | Allows direct measurement of adhesion forces between mature biofilms and antifouling surfaces [10]. |
| Vanillin-Modified Membranes | Anti-biofouling surfaces for testing the efficacy of biofilm adhesion reduction strategies. | Vanillin acts as a quorum-sensing inhibitor; used to validate novel force spectroscopy methods [10]. |
| Lawsone methyl ether | Lawsone methyl ether, CAS:2348-82-5, MF:C11H8O3, MW:188.18 g/mol | Chemical Reagent |
| BE-18591 | BE-18591, CAS:147138-01-0, MF:C22H35N3O, MW:357.5 g/mol | Chemical Reagent |
The advancement of High-Speed AFM as a tool for visualizing dynamic biofilm processes is directly linked to innovations in its core components. The development of novel cantilevers like the seesaw design overcomes fundamental SNR limitations, while advanced control algorithms such as DenseNet-type neural networks effectively linearize high-speed piezoelectric scanners. Together with sensitive optical detection systems, these components enable the capture of biofilm assembly and remodeling at unprecedented spatial and temporal resolutions. The protocols and materials detailed in this application note provide a foundation for researchers in drug development to implement and further refine HS-AFM techniques, ultimately contributing to the development of novel anti-biofilm strategies.
In the study of dynamic biofilm processes, understanding rapid initial attachment, cellular reorganization, and extracellular polymeric substance (EPS) deposition is crucial. These events occur on timescales of milliseconds to seconds, presenting a significant challenge for conventional imaging techniques. High-speed atomic force microscopy (HS-AFM) has emerged as a powerful tool capable of capturing these processes at sub-second resolution, providing unprecedented insight into biofilm development mechanics. This application note explores the fundamental physics enabling sub-second temporal resolution in HS-AFM, with specific emphasis on the high-speed feedback and tracking systems that make such imaging possible. The technical details presented herein are framed within the context of real-time biofilm research, providing practical methodologies for researchers investigating microbial community dynamics, antimicrobial efficacy, and biofilm-material interactions.
The Physics of High-Speed Feedback Systems
The achievement of sub-second resolution in HS-AFM is fundamentally governed by the feedback control system that maintains constant tip-sample interaction forces during high-speed scanning. The performance of this system is quantified by its feedback bandwidth (f_B), which represents the frequency at which the system can accurately track sample topography without significant phase lag. According to established HS-AFM principles, this bandwidth is mathematically expressed as:
fB = α / [8(τc + τa + τs + βτ_PID + δ)] [11]
Where the respective time constants correspond to different physical components of the system:
- Ï„c: Cantilever response time (Ï„c = Qc/(Ï€fc))
- Ï„_a: Deflection-to-amplitude converter response time
- Ï„s: Z-scanner response time (Ï„s = Qs/(Ï€fs))
- Ï„_PID: PID controller processing time
- δ: Sum of other minor time delays in the feedback loop
The correction factors α and β account for closed-loop operation and PID controller performance, respectively [11]. This equation demonstrates that achieving high feedback bandwidth requires minimizing each component's response time through specialized hardware and control strategies.
Key Instrumental Components for High-Speed Operation
Table 1: Critical HS-AFM Components and Their Performance Characteristics
| Component | Function | Performance Requirements | Impact on Speed |
|---|---|---|---|
| Short cantilevers | Measures tip-sample forces | High resonant frequency (fc ≈ 1.2 MHz in water), low spring constant (kc ≈ 0.15 N/m) [11] | Directly determines τ_c; higher fc reduces response time |
| High-speed Z-scanner | Positions sample vertically | High resonant frequency (fs), low quality factor (Qs) [11] | Minimizes Ï„_s for rapid vertical adjustments |
| Optical beam deflection sensor | Detects cantilever motion | High bandwidth, low noise | Enables fast detection of minute cantilever deflections |
| Fast D-to-A converter | Processes amplitude signals | High processing speed (n) relative to cantilever frequency [11] | Reduces Ï„_a for quicker signal conversion |
| Dynamic PID controller | Regulates feedback parameters | Gain-scheduling capability, minimal processing delay [11] | Optimizes Ï„_PID for varying surface features |
The integration of these specialized components enables modern HS-AFM systems to achieve imaging rates of 10-55 frames per second or higher, with temporal resolution reaching 10 microseconds and spatial resolution at the sub-nanometer level [12]. This performance is essential for capturing rapid structural changes in developing biofilms, such as flagellar coordination during initial surface attachment and the dynamics of EPS matrix formation.
Experimental Protocols for Biofilm Imaging
Protocol: Real-Time Visualization of Early Biofilm Formation
Objective: To capture the initial attachment phase of bacterial cells to surfaces with sub-second temporal resolution.
Materials:
- Pantoea sp. YR343 (gram-negative, rod-shaped bacterium with peritrichous flagella) [2]
- PFOTS-treated glass coverslips or silicon substrates [2]
- Appropriate liquid growth medium
- High-speed AFM system (e.g., Bruker NanoRacer) with small cantilevers (fc ≈ 1.2 MHz) [11] [12]
- Fluid cell for in-situ imaging
Methodology:
- Substrate Preparation: Treat glass coverslips with PFOTS to create a hydrophobic surface that promotes bacterial attachment while allowing for high-resolution AFM imaging [2].
- Sample Inoculation: Inoculate Petri dishes containing treated coverslips with Pantoea cells in liquid growth medium. Incubate for approximately 30 minutes to allow initial attachment.
- Sample Preparation: Gently rinse coverslips to remove unattached cells. For hydrated imaging, transfer immediately to AFM fluid cell. For higher resolution imaging of fixed structures, air-dry samples before imaging [2].
- HS-AFM Imaging:
- Mount sample in HS-AFM system equipped with appropriate small cantilevers
- Engage tapping mode in liquid with optimized free oscillation amplitude (Aâ‚€)
- Setpoint amplitude (A_s) should be approximately 90% of Aâ‚€ to minimize tip-sample forces while maintaining stability
- Implement gain-scheduling in PID controller to accommodate varying biofilm topography
- Acquire images at 10-50 frames per second depending on scan size
- Data Acquisition: Capture image sequences for 5-30 minutes to monitor initial attachment and microcolony formation events.
Expected Results: This protocol should yield high-resolution temporal sequences revealing individual bacterial cells (approximately 2 μm length × 1 μm diameter) with visible flagellar structures (20-50 nm height) interacting with the surface. Researchers should observe the distinctive honeycomb pattern formation that occurs during early biofilm development of Pantoea sp. YR343 [2].
Protocol: Line Scanning for Enhanced Temporal Resolution
Objective: To monitor rapid conformational changes in individual biofilm matrix components with millisecond resolution.
Materials:
- Mature biofilm samples (3-5 day old)
- HS-AFM system with line scanning capability
- Specialized cantilevers with high resonant frequencies (>1 MHz in liquid)
Methodology:
- Sample Preparation: Grow biofilms to maturity on appropriate substrates (3-5 days). Gently rinse with buffer to remove loosely attached cells.
- Region Selection: Using conventional HS-AFM imaging, identify areas of interest exhibiting dynamic structural features.
- Line Scanning Configuration:
- Position AFM tip over a single line crossing the feature of interest
- Reduce scan size to a single line (X-scan only, Y-position fixed)
- Maximize scan rate to achieve temporal resolution of 10-100 milliseconds
- Adjust feedback parameters to maintain stable tracking at high speeds
- Data Acquisition: Collect line scan data continuously for 1-5 minutes to capture transient events.
- Data Reconstruction: Utilize localization algorithms (e.g., NanoLocz) to extract sub-diffraction limit spatial information from temporal line scan data [12].
Expected Results: Line scanning enables observation of rapid structural dynamics in EPS components and membrane proteins with millisecond temporal resolution, revealing previously inaccessible information about matrix remodeling and molecular interactions within biofilms.
Figure 1: HS-AFM Feedback Control System. The diagram illustrates the critical components and their time constants within the high-speed feedback loop that enables sub-second resolution imaging. Minimizing each time constant (Ï„) is essential for achieving high feedback bandwidth.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 2: Key Research Reagent Solutions for HS-AFM Biofilm Studies
| Item | Function/Application | Specifications | Experimental Considerations |
|---|---|---|---|
| PFOTS-treated substrates | Hydrophobic surface for controlled bacterial attachment | (1H,1H,2H,2H-Perfluorooctyl)trichlorosilane treated glass or silicon | Promotes reproducible attachment while enabling high-resolution imaging [2] |
| Short cantilevers | High-speed force sensing | Length: 7 μm, Width: 2 μm, Thickness: 90 nm, Resonant frequency: ~1.2 MHz in water, Spring constant: ~0.15 N/m [11] | Essential for minimizing τ_c; commercially available from NanoAndMore, NanoWorld, RIBM |
| Pantoea sp. YR343 | Model biofilm-forming bacterium | Gram-negative, rod-shaped (2 μm length, 1 μm diameter), peritrichous flagella, forms honeycomb patterns [2] | Isolated from poplar rhizosphere; excellent model for studying early biofilm assembly |
| PEDOT-PSS sensors | Electrochemical impedance monitoring | Electrodeposited poly(3,4-ethylenedioxythiophene)-poly(styrenesulfonate) on Pt wire | Enables correlative impedance measurements during HS-AFM imaging [13] |
| NanoLocz software | Analysis of HS-AFM data | Automated leveling, particle detection, super-resolution algorithms [12] | Freely available tool for handling large HS-AFM datasets; enables localization AFM |
| Antcin B | Antcin B|3CLPro Inhibitor | Antcin B: SARS-CoV-2 3CLPro inhibitor for COVID-19 research. Also studies anticancer mechanisms. For Research Use Only. Not for human use. | Bench Chemicals |
| (-)-Hydroxycitric acid lactone | (-)-Hydroxycitric acid lactone, CAS:27750-10-3, MF:C6H8O8, MW:208.12 g/mol | Chemical Reagent | Bench Chemicals |
Figure 2: HS-AFM Biofilm Imaging Workflow. The experimental pathway from substrate preparation to high-temporal resolution imaging, highlighting critical steps where attention to technical details ensures successful capture of dynamic biofilm processes.
Applications in Biofilm Research
The implementation of HS-AFM with sub-second resolution has revealed previously inaccessible phenomena in biofilm development. Research utilizing the protocols outlined above has demonstrated:
Flagellar Coordination During Attachment: HS-AFM imaging of Pantoea sp. YR343 has revealed that flagellar interactions between adjacent cells exhibit coordinated patterns that facilitate the formation of distinctive honeycomb structures during early biofilm development [2].
Single-Molecule Dynamics in EPS: The combination of high temporal and spatial resolution has enabled researchers to visualize the assembly and disassembly of individual EPS components, providing insights into matrix remodeling dynamics in response to environmental stimuli [14].
Antimicrobial Mechanism Visualization: HS-AFM has captured the real-time interaction between antimicrobial peptides and bacterial membrane structures within biofilms, revealing pore formation dynamics and cellular response mechanisms at sub-second timescales [12].
These applications demonstrate how the physics of high-speed feedback and tracking enables researchers to move beyond static snapshots to truly dynamic understanding of biofilm processes, with significant implications for developing novel anti-biofilm strategies and optimizing biofilm-based biotechnological applications.
Atomic force microscopy (AFM) has become an indispensable tool for studying dynamic biological processes, such as biofilm formation and development, at the nanoscale. Its unique capability to characterize topography, adhesion, and nanomechanical properties in real-time and under physiologically relevant conditions provides insights unobtainable with other techniques [15] [2]. For biofilm research, understanding these parameters is crucial as they influence bacterial adhesion, community stability, resilience, and response to environmental challenges [16]. This application note details the key observable parameters and provides standardized protocols for their quantitative measurement using high-speed AFM modalities, enabling researchers to capture the dynamic nature of soft biological systems.
Key Parameters and Their Significance in Biofilm Research
The following parameters form the cornerstone of nanomechanical characterization in biofilm systems. Their quantitative assessment allows researchers to link structural features to functional outcomes in biofilm development and resistance mechanisms.
- Topography: AFM provides high-resolution, three-dimensional maps of surface morphology. For biofilms, this enables the visualization of individual cells, their arrangement within the community, and the structure of the extracellular polymeric substance (EPS) [2]. Topographical imaging has revealed features like the honeycomb pattern in early Pantoea sp. YR343 biofilms and the intricate network of flagella and other appendages that facilitate surface attachment and cell-cell communication [2].
- Adhesion: Adhesion forces measured by AFM reflect the chemical and physical interactions between the AFM tip (which can be functionalized) and the sample surface. In biofilms, this parameter is critical for understanding initial bacterial attachment to surfaces, cell-cohesion within the biofilm matrix, and the efficacy of anti-fouling surfaces [17] [16]. Mapping adhesion forces helps identify heterogeneous domains within the EPS that contribute to biofilm integrity.
- Young's Modulus (Elasticity): This quantitative measure of a material's stiffness is derived from force-distance curves. Biofilms are viscoelastic, and their elastic modulus is a key indicator of their mechanical strength and structural integrity [18] [16]. Monitoring changes in Young's modulus can reveal how biofilms respond to mechanical stress, nutrient availability, or antimicrobial agents.
- Deformation: The degree to which a sample is indented by the AFM tip under a given load. Softer materials, like many biofilms, exhibit higher deformation. This parameter is crucial for ensuring accurate modulus calculations and for understanding the local compliance of the biofilm, which can influence nutrient diffusion and cell protection [17].
- Energy Dissipation: This parameter quantifies the viscous energy loss during a loading-unloading cycle, providing a direct measure of the sample's viscoelasticity [18] [17]. For biofilms, a high dissipation value indicates a more liquid-like, dissipative behavior, which is a hallmark of their ability to absorb stress and resist mechanical removal [16].
Table 1: Key Nanomechanical Parameters and Their Significance in Biofilm Research
| Parameter | Description | Significance in Biofilm Research |
|---|---|---|
| Topography | 3D surface morphology and roughness | Reveals cellular arrangement, EPS structure, and biofilm architecture at the nanoscale [2]. |
| Adhesion Force | Maximum attractive force on tip retraction | Quantifies cell-surface and cell-cell interactions; predicts biofilm stability and adhesion strength [17]. |
| Young's Modulus | Quantitative measure of stiffness/elasticity | Indicates mechanical strength, structural integrity, and response to environmental challenges [18] [16]. |
| Deformation | Degree of sample indentation under load | Identifies local softness/compliance; crucial for accurate modulus calculation on soft samples [17]. |
| Energy Dissipation | Viscous energy loss during indentation | Measures viscoelasticity; predicts biofilm's ability to absorb energy and resist mechanical disruption [17] [16]. |
High-Speed AFM Modes for Real-Time Measurement
Conventional AFM modes can be too slow to capture dynamic processes. The following advanced modes significantly increase data acquisition rates, enabling real-time observation of biofilm development.
- PinPoint Nanomechanical Mode: This mode operates by stopping the XY scanner at each pixel and acquiring a high-speed force-distance curve. The decoupled motion of the Z scanner eliminates lateral shear forces, minimizing sample damage [17]. Its primary advantage is the simultaneous, real-time acquisition of topography, modulus, adhesion, deformation, and dissipation images with high signal-to-noise ratio. This makes it ideal for quantifying mechanical heterogeneity in static or slowly evolving biofilm structures.
- High-Speed AFM (HS-AFM): HS-AFM utilizes miniaturized cantilevers and high-frequency feedback systems to achieve video-rate imaging (sub-second frame acquisition) [8]. This is the preferred technique for directly visualizing dynamic events, such as the movement of individual membrane proteins, the growth of EPS fibrils, or the real-time interaction between bacterial cells within a developing biofilm [8].
- Bimodal AFM: This multifrequency technique excites and measures the cantilever's response at two eigenmodes (resonant frequencies) simultaneously. It provides enhanced material contrast beyond topography by sensitively mapping variations in nanomechanical properties [19]. By incorporating nonlinear response at harmonics and mixing frequencies, material discrimination can be improved almost threefold, allowing for exquisite differentiation of components within the complex biofilm matrix without sacrificing imaging speed [19].
Table 2: Comparison of High-Speed AFM Modes for Biofilm Characterization
| AFM Mode | Key Principle | Typical Resolution | Data Output | Best for Biofilm Applications |
|---|---|---|---|---|
| PinPoint Nanomechanical | Sequential high-speed force-distance curves [17] | Nanometer lateral, piconewton force | Quantitative maps of modulus, adhesion, deformation [17] | Quantifying static mechanical heterogeneity; correlating structure with function. |
| High-Speed AFM (HS-AFM) | Miniaturized cantilevers with fast feedback [8] | Molecular to cellular scale | Real-time video of topographical changes [8] | Imaging dynamic processes: protein diffusion, appendage movement, initial attachment. |
| Bimodal AFM | Dual-eigenmode excitation with nonlinear analysis [19] | Nanometer lateral, high material contrast | Topography and qualitative/quantitative property maps [19] | High-speed mapping of material components within the EPS with superior contrast. |
Experimental Protocols
Protocol 1: Sample Preparation for Biofilm AFM
Proper sample preparation is critical for obtaining reproducible nanomechanical data that reflects the native state of the biofilm [15].
1. Substrate Selection and Functionalization:
- Substrates: Use atomically flat substrates such as freshly cleaved mica, silicon wafers, or glass coverslips to minimize background roughness [15].
- Functionalization: To promote bacterial adhesion, treat the substrate. For example, coat mica with poly-lysine or glass with polyethyleneimide (PEI) to introduce a positive charge for electrostatic interaction with negatively charged cell surfaces [15].
2. Biofilm Growth and Harvesting:
- Culture Conditions: Grow biofilms under controlled, relevant conditions (e.g., flow cell, agar plate, or liquid culture) to the desired developmental stage.
- Inoculation: For time-course studies, inoculate the functionalized substrate with a bacterial suspension (e.g., Pantoea sp. YR343) and incubate for specific periods (e.g., 30 minutes for initial attachment, 6-8 hours for microcolony formation) [2].
- Rinsing: Gently rinse the substrate with a appropriate buffer (e.g., PBS) to remove non-adherent planktonic cells. Avoid harsh rinsing that could disrupt delicate structures like flagella [2].
3. Mounting for AFM Imaging:
- Liquid vs. Air Imaging: For high-resolution imaging and faithful mechanical characterization, perform AFM in liquid using a fluid cell. This preserves the native hydration state and physiology of the biofilm [2] [16]. If imaging in air is necessary, a brief, careful air-drying step may be used, but this can alter nanomechanical properties and introduce artifacts.
Protocol 2: Quantitative Nanomechanical Mapping
This protocol outlines the steps for performing nanomechanical mapping on a biofilm sample using a mode like PinPoint.
1. Cantilever Selection and Calibration:
- Selection: Use soft cantilevers (spring constant 0.1 - 1 N/m) for soft biological samples like biofilms to ensure sufficient deformation without damaging the sample [17]. The tip geometry (e.g., spherical tip) can help avoid sample piercing.
- Calibration: Precisely calibrate the cantilever's spring constant (using thermal tune or Sader method) and the optical lever sensitivity on a hard, non-deformable reference sample (e.g., sapphire) before measurement [15] [17]. This is non-negotiable for quantitative modulus values.
2. Measurement Parameter Optimization:
- Setpoint Force: Use the lowest possible setpoint force that maintains stable tip-sample contact. High forces will cause excessive indentation and potentially damage the delicate biofilm structure [15].
- Approach/Retract Speed: Optimize the Z-scan rate. Too fast can cause hydrodynamic effects and miss true contact point; too slow limits throughput. For PinPoint modes, high speeds (e.g., 10x conventional force-volume) are achievable [17].
- Spatial Resolution: Set the pixel resolution (e.g., 256 x 256 or 512 x 512) based on the feature size and required field of view. Balance between resolution and total acquisition time to minimize drift.
3. Data Acquisition and Model Fitting:
- Acquisition: Acquire maps over regions of interest (e.g., a single cell, a cell cluster). Ensure the sample is thick enough (>10x the indentation depth) to prevent substrate effects [15].
- Model Selection: Fit the retraction portion of the force-distance curves with an appropriate contact mechanics model [17]:
- Hertz model: For non-adhesive elastic contact.
- DMT model: For adhesive elastic contact on stiffer samples.
- JKR model: For adhesive elastic contact on softer samples (most applicable for biofilms).
- Analysis: Use the instrument's software (e.g., SmartScan) or custom scripts (e.g., in Python) to automatically process all force-curves in a map and generate spatially correlated maps of Young's modulus, adhesion, and deformation [17].
The Scientist's Toolkit
Table 3: Essential Research Reagent Solutions for Biofilm AFM
| Item | Function/Description | Example Use Case |
|---|---|---|
| Soft Cantilevers | Probes with low spring constant (0.1 - 1 N/m); essential for soft sample indentation without damage. | Nanomechanical mapping of live bacterial cells and soft EPS [17]. |
| Functionalized Substrates | Chemically modified surfaces (e.g., poly-lysine coated mica) to promote controlled bacterial adhesion. | Studying initial stages of bacterial attachment and biofilm formation [15] [2]. |
| Calibration Samples | Hard, non-deformable samples (e.g., sapphire) and grating samples for accurate probe calibration. | Calibrating cantilever sensitivity and verifying scanner accuracy for quantitative measurements [17]. |
| Liquid Cell | A sealed chamber that allows the AFM tip and sample to be immersed in buffer or growth medium. | Imaging biofilms in physiologically relevant liquid environments [2] [16]. |
| Biofilm Growth Media | Nutritional solutions tailored to the specific bacterial strain to support robust biofilm development. | Culturing mature, native-state biofilms for mechanical testing [2]. |
| O-Acetylcamptothecin | Camptothecin, Acetate|Research Grade Topoisomerase I Inhibitor | Camptothecin, acetate is a research compound for studying Topoisomerase I inhibition and cancer mechanisms. For Research Use Only. Not for human or veterinary use. |
| Hinokinin | Hinokinin|Lignan|For Research Use Only |
From Theory to Practice: Methodological Guide for HS-AFM in Biofilm Research
Within the broader context of high-speed atomic force microscopy (AFM) research on dynamic biofilm processes, obtaining stable, high-resolution images presents a significant challenge. Biofilms are structurally complex, heterogeneous microbial communities encased in extracellular polymeric substances (EPS) [2] [20]. While AFM offers unparalleled nanoscale resolution for imaging live cells under physiological conditions, its application is critically dependent on effective sample preparation [21] [22]. The lateral forces exerted by the scanning AFM tip can easily displace weakly adhered cells, compromising image quality and biological relevance [23]. Therefore, the immobilization protocol is not merely a preliminary step but a cornerstone for successful experimentation, enabling researchers to investigate dynamic events such as initial attachment, cellular division, and response to antimicrobial agents without introducing artifacts that distort true biofilm physiology. This document details standardized protocols for immobilizing live biofilms, specifically tailored for high-speed AFM imaging.
Immobilization Method Selection and Comparison
Choosing an appropriate immobilization strategy is a trade-off between achieving mechanical stability and preserving native cell viability and function. The optimal method depends on the bacterial strain, the scientific questions being addressed, and the desired imaging conditions. The table below provides a comparative overview of three established approaches.
Table 1: Comparison of Bacterial Immobilization Methods for AFM Imaging
| Method | Mechanism of Immobilization | Best Suited For | Key Advantages | Key Limitations |
|---|---|---|---|---|
| Gelatin-Coated Mica [22] | Electrostatic interaction between negatively charged bacteria and a positively charged gelatin film. | Gram-negative and Gram-positive bacteria; high-resolution imaging in liquid; force spectroscopy. | Minimally invasive; preserves cell viability and division; suitable for dynamic studies. | Efficiency depends on gelatin source (porcine recommended); susceptible to interference from growth media and salts. |
| Poly-L-Lysine (PLL) Coated Surfaces [21] | Cationic polymer binds negatively charged bacterial cells and surface. | Strains with low inherent adhesiveness; imaging in various ionic strength buffers. | Strong, reliable adhesion in both low and high ionic strength buffers. | Can compromise cell membrane integrity if not optimized; requires addition of divalent cations for viability. |
| Immobilization-Free [23] | Uses fast force-distance curve-based AFM mode (e.g., QI mode) to eliminate lateral forces. | Motile bacteria (e.g., gliding strains); studies where any surface chemistry is undesirable. | No chemical or mechanical stress from immobilization; ideal for observing native gliding motility. | Requires specialized AFM operation modes; not all commercial systems may be compatible. |
Detailed Experimental Protocols
Protocol 1: Immobilization on Gelatin-Coated Mica
This protocol is a robust and widely applicable method for imaging a broad range of bacterial strains in liquid conditions [22].
The Scientist's Toolkit: Key Reagent Solutions
- Gelatin (Sigma G-6144 or G-2625): A porcine-derived protein that forms a positively charged coating on mica, facilitating electrostatic immobilization of cells [22].
- Freshly Cleaved Mica: An atomically flat, negatively charged substrate that provides an ideal surface for the gelatin coating and subsequent imaging [22].
- Divalent Cations (Mg²âº, Ca²âº): Often added to immobilization or imaging buffers to help stabilize bacterial cell membranes and enhance viability [21].
- Dilute Phosphate Buffered Saline (PBS) or Sucrose Solution: Used as an imaging buffer. Low ionic strength is crucial for stable immobilization, and sucrose can be used to maintain osmotic balance for sensitive cells [22].
Procedure:
- Mica Preparation: Cut mica to fit the AFM sample disk (approximately 22 x 30 mm). Cleave the mica on both sides using adhesive tape to expose a fresh, smooth surface [22].
- Gelatin Solution Preparation: Add 0.5 grams of porcine gelatin to 100 mL of boiling distilled water. Gently swirl until the gelatin is completely dissolved. Cool the solution to 60-70 °C before use [22].
- Mica Coating: Submerge the cleaved mica square into the warmed gelatin solution and withdraw it quickly. Place the coated mica on edge on a paper towel to dry in ambient air. The coated mica can be stored and used for up to two weeks [22].
- Bacterial Sample Preparation: Pellet 1 mL of a bacterial culture (OD₆₀₀ ≈ 0.5 - 1.0) by centrifugation. Wash the pellet in a filtered, low-ionic-strength buffer (e.g., deionized water or dilute PBS) to remove growth media and salts that can interfere with adhesion. Resuspend the pellet in 500 µL of the same buffer to create a turbid suspension [22].
- Cell Deposition: Apply 10-20 µL of the bacterial suspension to the gelatin-coated mica. Spread the droplet gently with a pipette tip without touching the surface. Allow the sample to incubate for 10 minutes [22].
- Rinsing: Gently rinse the mica surface with a stream of distilled water or imaging buffer to remove loosely bound cells. The success of immobilization can be tested by allowing the sample to dry; a cloudy area indicates successful cell adhesion. For live imaging, keep the sample hydrated [22].
- AFM Imaging: Mount the sample in the AFM liquid cell and image in a compatible liquid medium such as 0.005 M PBS or a sucrose solution. Use non-contact imaging modes (e.g., Tapping Mode) or contact mode with low spring constant cantilevers to minimize lateral forces [22].
Protocol 2: Immobilization on Poly-L-Lysine Surfaces
This method provides stronger adhesion, which is useful for less adherent strains, but requires careful optimization to maintain cell viability [21].
Procedure:
- Surface Coating: Prepare a 0.1% (w/v) aqueous solution of poly-L-lysine (PLL). Apply a few drops to a clean glass or mica substrate for 5-10 minutes. Remove the excess solution and allow the surface to air dry.
- Optimized Immobilization Buffer Preparation: Use a lower ionic strength buffer supplemented with divalent cations (e.g., Mg²⺠and Ca²⺠at ~1-5 mM) and glucose to mitigate the detrimental effects of PLL on membrane integrity [21].
- Bacterial Preparation and Deposition: Prepare a bacterial pellet as in Protocol 1, but resuspend in the optimized immobilization buffer. Apply the cell suspension to the PLL-coated surface and allow it to settle for a defined period (e.g., 10-30 minutes).
- Rinsing and Imaging: Gently rinse with the immobilization buffer to remove non-adherent cells. Proceed with AFM imaging in a compatible buffer. It is critical to monitor cell viability throughout the process using membrane integrity assays [21].
Workflow for Method Selection
The following diagram outlines a logical decision pathway for selecting and applying the most appropriate immobilization protocol based on research objectives and bacterial characteristics.
Advanced Applications and Future Directions
The field of biofilm immobilization is evolving with AFM technology. For studying highly dynamic processes like the gliding motility of filamentous cyanobacteria, immobilization-free techniques are groundbreaking. These methods utilize advanced AFM modes that acquire fast force-distance curves at every pixel, drastically reducing lateral forces and eliminating the need for chemical or physical entrapment [23]. This allows for the in-situ determination of mechanical properties like Young's modulus and turgor pressure while observing native bacterial movement [23].
Furthermore, automation and machine learning (ML) are beginning to transform sample preparation and imaging. Automated large-area AFM systems can now perform high-resolution imaging over millimeter-scale areas, overcoming the limited scan range of traditional AFMs [2]. ML algorithms are crucial for stitching these images together and for the automated segmentation, detection, and classification of cells within complex biofilm architectures, enabling quantitative analysis of parameters like cell count, confluency, and orientation over large, representative areas [2]. These advancements, combined with robust immobilization protocols, are pushing the boundaries of high-speed AFM from single-cell observations to the study of entire biofilm community dynamics.
Atomic force microscopy (AFM) is a powerful tool for high-resolution imaging of biological samples, including biofilms, under physiological conditions. However, conventional AFM is limited by a fundamental scale mismatch: its scan range is typically restricted to areas less than 100 micrometers, while biofilms and other complex microbial communities form heterogeneous structures spanning millimeters or more [2]. This limitation has historically obstructed a comprehensive understanding of how local cellular and sub-cellular interactions give rise to functional macroscale organization.
Automated large-area AFM represents a transformative solution to this problem. This approach integrates hardware automation, advanced image stitching algorithms, and machine learning to seamlessly combine hundreds or thousands of high-resolution scans into a single, coherent millimeter-scale image [2] [24]. For researchers investigating dynamic biofilm processes, this technology enables the correlation of nanoscale features—such as individual appendages and extracellular polymeric substances (EPS)—with emergent community-scale patterns and heterogeneities that dictate biofilm function and resilience.
Key Technological Advancements
Core Hardware and Software Infrastructure
The implementation of automated large-area AFM rests on two foundational pillars: programmatic control of the AFM hardware and intelligent image reconstruction software.
- Automated Operation: Traditional AFM operation is labor-intensive and requires specialized operators, making large-scale surveys impractical. The development of sophisticated Application Programming Interfaces (APIs), such as the Python library provided by Nanosurf, enables full scripting control over the AFM [24]. This automation allows for pre-programmed raster scanning patterns over centimeter-scale areas with minimal user intervention, facilitating continuous, multi-day experiments [2].
- Seamless Image Stitching: Acquiring a large-area dataset generates hundreds of individual image tiles. A critical challenge is stitching these tiles together accurately despite minimal overlap, which maximizes acquisition speed. Advanced computational algorithms merge these tiles by identifying matching features along the edges, creating a seamless, high-resolution map of the entire scanned area [2].
The Role of Machine Learning and Artificial Intelligence
Machine learning (ML) and artificial intelligence (AI) are integral to making large-area AFM efficient, quantitative, and autonomous. Their applications span the entire experimental workflow [2].
- Scanning Process Optimization: ML models can refine tip-sample interactions, correct for image distortions, and employ sparse scanning approaches to significantly reduce data acquisition time [2].
- Autonomous Operation: AI-driven systems can automate routine tasks such as probe conditioning and approach. In advanced setups, large language models have even been used for direct control of the AFM, enabling truly autonomous experimentation [2].
- Image Analysis and Quantification: The massive datasets generated by large-area AFM necessitate automated analysis. ML algorithms excel at segmenting images, detecting individual cells, and classifying morphological features without human bias. This allows for the efficient extraction of quantitative parameters such as cell count, confluency, shape, and orientation across the entire millimeter-scale map [2].
Table 1: Machine Learning Applications in Automated Large-Area AFM
| Application Area | Specific Function | Benefit |
|---|---|---|
| Data Acquisition | Optimizes scanning site selection & tip conditioning [2] | Reduces human intervention, increases speed |
| Scanning Process | Corrects distortions, enables sparse scanning [2] | Enhances data quality and acquisition efficiency |
| Image Analysis | Automated cell detection, segmentation, and classification [2] | Enables quantitative analysis of massive datasets |
| Autonomous Control | Direct control via AI and language models [2] | Allows for continuous, multi-day experiments |
Experimental Protocols
Protocol: Large-Area AFM of Early-Stage Biofilm Formation
This protocol outlines the procedure for studying the initial attachment and organization of bacterial cells on a surface using automated large-area AFM, as applied to Pantoea sp. YR343 [2].
Materials and Reagents
- Bacterial Strain: Pantoea sp. YR343 (or other relevant strain) growing in a liquid growth medium [2].
- Substrate: Glass coverslips treated with (Heptadecafluoro-1,1,2,2-tetrahydrooctyl)trichlorosilane (PFOTS) to create a hydrophobic surface [2].
- Equipment: Atomic Force Microscope equipped with a large-area scanner and automated stage. A cantilever suitable for tapping mode in air or liquid.
Procedure
Sample Preparation:
- Place the PFOTS-treated glass coverslips in a petri dish.
- Inoculate the dish with the Pantoea sp. YR343 culture in liquid growth medium.
- Incubate for a designated initial attachment period (e.g., 30 minutes).
- At the selected time point, carefully remove a coverslip and gently rinse it with a buffer solution (e.g., deionized water) to remove any non-adherent cells.
- Air-dry the sample before imaging. While AFM can be performed in liquid, the cited study involved dried samples for high-resolution imaging of appendages [2].
AFM Setup and Large-Area Scan Configuration:
- Mount the prepared sample onto the AFM stage.
- Select a cantilever and engage it onto the surface in a representative area.
- Using the scripting interface (e.g., Python API), define the large-area scan parameters:
- Total Scan Area: Define the millimeter-scale region of interest (e.g., 1 mm x 1 mm).
- Individual Tile Size: Set the size of each high-resolution image (e.g., 50 µm x 50 µm).
- Tile Overlap: Specify a minimal overlap (e.g., 5-10%) between adjacent tiles to facilitate stitching.
Automated Data Acquisition:
- Initiate the automated scanning script. The system will sequentially acquire high-resolution AFM images for each predefined tile across the entire sample area.
- The process may run for several hours automatically, depending on the size of the area and the resolution of individual scans.
Post-Processing and Image Analysis:
- Image Stitching: Use the dedicated software algorithm to merge all individual image tiles into a single, large-area topographic map.
- Machine Learning Analysis: Apply trained ML models to the stitched image to automatically:
- Identify and segment individual bacterial cells.
- Classify cells based on morphology.
- Quantify parameters such as cell density, distribution, and orientation.
- Detect and analyze nanoscale features like flagella [2].
Expected Results
- The final stitched image will reveal the spatial organization of surface-attached cells over a millimeter-scale area.
- For Pantoea sp. YR343, analysis should uncover a preferred cellular orientation and the emergence of a distinctive honeycomb pattern in cell clusters after 6-8 hours of growth [2].
- High-resolution segments of the large-area map will allow for the visualization of flagella and other appendages, suggesting their role in coordinating biofilm assembly beyond initial attachment [2].
Protocol: Combinatorial Screening of Surface Modifications
This methodology leverages large-area AFM to conduct high-throughput analysis of how different surface properties influence bacterial adhesion and biofilm formation [2].
Materials and Reagents
- Gradient Surfaces: Silicon substrates with engineered surface property gradients (e.g., chemical composition, topography, or stiffness).
- Bacterial Strain: A relevant bacterial strain of interest.
Procedure
Sample Preparation:
- Inoculate the gradient-structured surface with the bacterial culture.
- Incubate for a set time to allow for initial attachment.
- Rinse gently and air-dry, as described in Protocol 3.1.2.
AFM Imaging Across the Gradient:
- Mount the gradient sample.
- Program the automated AFM to perform a series of large-area scans at predefined intervals along the surface gradient.
- This generates multiple, large-area maps, each corresponding to a different set of surface properties.
Quantitative Comparison:
- For each large-area map, use ML-based analysis to calculate the bacterial density and morphological data.
- Correlate these quantitative metrics with the known surface properties at each location.
Expected Results
- This approach will identify specific surface modifications that significantly reduce bacterial density [2].
- The data provides a combinatorial understanding of how surface properties control adhesion, offering a powerful strategy for developing anti-biofilm surfaces.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for Large-Area AFM Biofilm Studies
| Item | Function/Description | Example/Reference |
|---|---|---|
| Functionalized Substrates | Engineered surfaces to study adhesion; PFOTS creates a hydrophobic surface for attachment studies [2]. | PFOTS-treated glass [2] |
| Automated AFM Platform | AFM system with a large-range scanner and a scripting API for automated control. | Nanosurf platforms with Python API [24] |
| Image Stitching Software | Computational tool to merge hundreds of high-res AFM tiles into a seamless large-area map. | Custom algorithms with minimal feature matching [2] |
| Cell Detection ML Model | Machine learning algorithm for automated identification and segmentation of cells in large datasets. | Convolutional Neural Networks (CNN) for image segmentation [2] |
| High-Resolution AFM Probes | Sharp cantilevers essential for resolving nanoscale features like flagella and pili. | Cantilevers for tapping mode in air or liquid |
| Paclobutrazol | Paclobutrazol, CAS:66346-05-2, MF:C15H20ClN3O, MW:293.79 g/mol | Chemical Reagent |
| 2',4'-Dihydroxyacetophenone | 2',4'-Dihydroxyacetophenone, CAS:5706-85-4, MF:C8H8O3, MW:152.15 g/mol | Chemical Reagent |
Workflow and Data Interpretation
The following diagram illustrates the integrated workflow of automated large-area AFM, from sample preparation to quantitative insight.
Automated large-area AFM represents a paradigm shift in high-speed AFM imaging for dynamic biofilm research. By overcoming the critical spatial limitation of conventional AFM, this technology directly links nanoscale structural and functional properties to the emergent macroscale organization of biofilms. The integration of full hardware automation with machine learning for both image reconstruction and quantitative analysis transforms AFM from a manual, single-spot characterization tool into a powerful, high-throughput platform for surveying biological complexity across its inherent length scales. This capability is crucial for advancing our understanding of biofilm development, heterogeneity, and response to environmental stresses, ultimately accelerating the development of effective anti-biofilm strategies in therapeutic and industrial contexts.
The following tables summarize key quantitative parameters from high-speed Atomic Force Microscopy (HS-AFM) studies on biofilm formation, focusing on the early attachment and structural development of Pantoea sp. YR343.
Table 1: HS-AFM Imaging Parameters for Biofilm Analysis
| Parameter | Specification | Application Note |
|---|---|---|
| Imaging Area | Up to millimeter-scale | Enables statistical analysis by capturing numerous cells and clusters in a single scan [2]. |
| Spatial Resolution | Nanoscale (sub-50 nm for flagella) | Resolves fine features like flagella (~20-50 nm height) and extracellular polymeric substances (EPS) [2]. |
| Z-axis Resolution | ~0.10 nm (with 12-bit capture) | Provides high vertical sensitivity for detailed topographical mapping [25]. |
| Digital Resolution | 12-bit (4096 values) or higher | Preserves data integrity and prevents information loss during processing and export [25]. |
| Scan Pattern | Zig-zag raster (typical) | Standard method for data acquisition; overscan regions (~30%) are often used in high-speed modes [25]. |
Table 2: Quantitative Morphological Data from Early Pantoea sp. YR343 Biofilm Development
| Biofilm Stage | Key Observations | Quantitative Measurements | Significance |
|---|---|---|---|
| Initial Attachment (~30 min) | Single, dispersed cells with visible flagella and pili [2]. | Cell length: ~2 µm; Diameter: ~1 µm; Surface area: ~2 µm² [2]. | Reveals the first point of surface contact and the role of appendages [2]. |
| Microcolony Formation (6-8 hours) | Cell clusters forming a distinctive "honeycomb" pattern with characteristic gaps [2]. | Flagella observed bridging gaps between cells, extending tens of micrometers [2]. | Suggests flagellar coordination is crucial beyond initial attachment, aiding in cluster organization [2]. |
| Spatial Heterogeneity | Automated large-area AFM maps reveal preferred cellular orientation and pattern formation [2]. | High-throughput analysis of cell count, confluency, shape, and orientation over millimeter areas [2]. | Links nanoscale cellular features to the functional macroscale organization of the emerging biofilm [2]. |
Experimental Protocols
Protocol: Sample Preparation for HS-AFM of Bacterial Initial Attachment
This protocol details the procedure for preparing bacterial samples on solid substrates to visualize the early stages of biofilm formation using HS-AFM [2].
Key Reagents & Materials:
- Bacterial Strain: Pantoea sp. YR343 (or other relevant strain) [2].
- Substrate: Glass coverslips, treated with PFOTS ((Heptadecafluoro-1,1,2,2-tetrahydrooctyl)trichlorosilane) or other functionalizing agents [2].
- Growth Medium: Appropriate liquid growth medium (e.g., Lysogeny Broth) [2].
- Equipment: Sterile Petri dishes, inoculation loops, pipettes, biosafety cabinet.
Procedure:
- Substrate Preparation: Place a PFOTS-treated glass coverslip into a sterile Petri dish. PFOTS treatment creates a hydrophobic surface that promotes specific bacterial attachment [2].
- Inoculation: Inoculate the Petri dish with Pantoea cells suspended in a liquid growth medium [2].
- Incubation: Allow the sample to incubate at the appropriate temperature (e.g., 28-30°C for Pantoea) for a selected time (e.g., 30 minutes for initial attachment studies) [2].
- Rinsing: At the designated time point, carefully remove the coverslip from the Petri dish and gently rinse it with a buffer solution (e.g., deionized water or PBS) to remove any non-adherent (planktonic) cells [2].
- Drying: Air-dry the coverslip at ambient temperature before mounting it on the AFM sample stage. Note: While AFM can be performed in liquids, this specific protocol involved drying to preserve nanoscale features like flagella for high-resolution imaging [2].
- Imaging: Mount the prepared sample on the AFM for large-area, high-resolution scanning.
Protocol: Automated Large-Area HS-AFM for Biofilm Structural Analysis
This protocol describes the operation of an automated large-area HS-AFM system to capture the spatial heterogeneity of a developing biofilm [2].
Key Reagents & Materials:
- Prepared bacterial sample on a substrate (from Protocol 2.1).
- AFM System: Equipped with a large-range scanner (capable of millimeter-scale travel).
- AFM Probes: Sharp silicon nitride or silicon tips appropriate for contact or tapping mode in air or liquid.
- Computer with automated AFM control and machine learning (ML) image analysis software.
Procedure:
- System Calibration: Calibrate the AFM's piezoelectric scanners in the X, Y, and Z axes using a standard calibration grating with known feature sizes and heights [25].
- Initial Scan Region Selection: Manually or using an ML-assisted overview scan, identify a representative region of interest on the sample surface to begin automated imaging [2].
- Automated Large-Area Scanning:
- Configure the software to automatically acquire multiple contiguous high-resolution AFM images (tiles) across a millimeter-sized area [2].
- Set imaging parameters (e.g., scan speed, resolution per tile, set-point) to optimize for biological samples, balancing speed and resolution to avoid sample damage.
- The system will sequentially scan and save each tile with minimal overlap to maximize acquisition speed [2].
- Image Stitching: Use integrated machine learning algorithms to seamlessly stitch the individual image tiles into a single, high-resolution, large-area map with minimal matching features [2].
- Data Processing:
- Perform initial leveling (plane/line fitting) using open-source software (e.g., Gwyddion) or custom scripts (e.g., TopoStats) to remove sample tilt and scan line noise [25].
- Apply non-destructive filters (e.g., Gaussian filter) if needed for noise reduction and enhanced visualization, ensuring quantitative measurements are performed on the raw, leveled data [25].
- Automated Image Analysis:
Research Reagent Solutions
Table 3: Essential Research Reagents and Materials for HS-AFM Biofilm Studies
| Item | Function/Application in HS-AFM Biofilm Research |
|---|---|
| PFOTS-Treated Substrates | Creates a defined hydrophobic surface chemistry to study the effects of surface properties on initial bacterial adhesion and subsequent biofilm architecture [2]. |
| Functionalized AFM Probes | Sharp tips (silicon nitride) are used for high-resolution topographical imaging. Tips can be functionalized with specific chemicals or biomolecules to measure interaction forces or map chemical properties [2]. |
| Open-Source Analysis Software (Gwyddion, TopoStats) | Provides standardized, and often automated, processing and analysis of AFM data, including leveling, particle analysis, and quantitative extraction of morphological and mechanical properties [25]. |
| Machine Learning Segmentation Tools | Integrated into the AFM data pipeline to automate the detection, counting, and classification of thousands of cells from large-area scans, enabling robust statistical analysis [2]. |
| Localization AFM (LAFM) Software | A post-acquisition image analysis method that uses spatial fluctuations in topographic features to achieve sub-nanometer resolution (~4 Ã…), revealing unprecedented structural details on protein surfaces within biofilms [25]. |
Workflow and Data Processing Visualization
Diagram 1: Integrated workflow for automated large-area HS-AFM analysis of biofilms, from sample preparation to quantitative data output.
Diagram 2: Step-by-step data processing pipeline for HS-AFM images, emphasizing the preservation of digital resolution and quantitative integrity.
Integrating Machine Learning for Automated Cell Detection, Classification, and Data Analysis
Application Notes
The study of dynamic biofilm processes requires analytical techniques that can resolve structural heterogeneity and temporal changes across multiple spatial scales. Atomic force microscopy (AFM) provides high-resolution topographical, mechanical, and functional insights at the nanoscale, yet its conventional imaging area is typically limited to less than 100 µm, creating a scale mismatch with the millimeter-scale organization of microbial communities [2]. This application note details the integration of machine learning (ML) with an automated large-area AFM approach to overcome this limitation, enabling comprehensive analysis of biofilm assembly, from individual cellular features to emergent community architecture. This methodology is framed within broader thesis research aimed at understanding the fundamental principles governing biofilm development and resilience, with direct implications for combating biofilm-associated infections and designing anti-fouling surfaces in clinical and industrial settings [2] [26].
Key Experimental Findings and Quantitative Data
The implemented automated large-area AFM system, combined with ML-based analysis, has been successfully applied to investigate the early attachment and biofilm formation of Pantoea sp. YR343. The system captures high-resolution images over millimeter-scale areas, revealing previously obscured spatial patterns and cellular interactions [2].
Table 1: Summary of Key Experimental Findings from Large-Area AFM Biofilm Imaging
| Parameter | Finding | Significance |
|---|---|---|
| Preferred Cellular Orientation | Distinctive honeycomb pattern formation [2] | Suggests coordinated cell-cell interactions during early biofilm assembly. |
| Flagellar Interactions | Flagellar structures (~20–50 nm height) bridging cellular gaps [2] | Indicates a role for flagella in biofilm assembly beyond initial surface attachment. |
| Single-Cell Morphology | Rod-shaped cells ~2 µm in length and ~1 µm in diameter [2] | Provides a baseline for identifying morphological changes in mutants or under stress. |
| ML Classification Accuracy (Biofilms) | Mean accuracy of 0.66 ± 0.06 for classifying staphylococcal biofilm maturity [26] | Demonstrates the feasibility of automated, objective assessment of biofilm development stages. |
| Human Classification Accuracy (Biofilms) | Mean accuracy of 0.77 ± 0.18 for the same task [26] | Highlights the performance of ML as comparable to, though slightly less accurate than, human experts. |
The application of this integrated system allows for the quantitative extraction of parameters such as cell count, confluency, cell shape, and orientation over large areas, directly linking nanoscale cellular appendages like flagella to the development of the microscale honeycomb pattern [2].
Experimental Protocols
Protocol 1: Large-Area AFM Imaging of Early Biofilm Formation
Application: This protocol describes the procedure for preparing samples and using an automated large-area AFM to image the early stages of bacterial attachment and biofilm formation on a surface [2].
Primary Model System: Pantoea sp. YR343 (a gram-negative, rod-shaped bacterium with peritrichous flagella) on PFOTS-treated glass coverslips [2].
Reagents and Materials:
- Pantoea sp. YR343 culture in appropriate liquid growth medium.
- Sterile Petri dishes.
- PFOTS-treated glass coverslips.
- Relevant buffers for gentle rinsing.
Procedure:
- Inoculation: Place PFOTS-treated glass coverslips in a sterile Petri dish and inoculate with Pantoea cells suspended in liquid growth medium.
- Incubation: Incubate the Petri dish for a selected time (e.g., ~30 minutes for initial attachment studies; 6-8 hours for early cluster formation) under suitable environmental conditions.
- Sample Harvesting: At the desired time point, carefully remove a coverslip from the Petri dish.
- Rinsing: Gently rinse the coverslip with a suitable buffer to remove non-adherent or loosely attached cells.
- Drying: Allow the sample to dry before AFM imaging.
- AFM Imaging: Mount the sample on the AFM stage. Use the automated large-area scanning routine to capture multiple contiguous high-resolution images over millimeter-scale areas. The automation includes predefined scanning coordinates and minimal overlap between adjacent images to maximize acquisition speed.
Protocol 2: Machine Learning-Assisted Classification of Biofilm Maturity
Application: This protocol outlines the steps for acquiring AFM images and using a dedicated machine learning algorithm to classify the maturity stage of staphylococcal biofilms based on topographic characteristics [26].
Primary Model System: Staphylococcus aureus biofilms.
Reagents and Materials:
- Relevant culture medium and reagents for growing S. aureus biofilms.
- Substrates for biofilm growth (e.g., glass, plastic).
- Atomic Force Microscope.
Procedure:
- Biofilm Growth: Grow S. aureus biofilms on appropriate substrates for varying durations to obtain a range of maturity stages.
- AFM Imaging: Image the biofilms using AFM to capture topographical data. The images should reveal characteristics related to the substrate, bacterial cells, and extracellular matrix.
- Ground Truth Establishment: A set of AFM images should be classified by human researchers into pre-defined maturity classes (e.g., a scheme of 6 classes) to establish a ground truth dataset.
- ML Algorithm Application: Process the AFM images using the developed deep learning algorithm. The algorithm is designed to identify pre-set topographic characteristics and assign a maturity class.
- Validation and Analysis: Compare the algorithm's classification results against the established ground truth. Calculate performance metrics such as accuracy, recall, and off-by-one accuracy (the proportion of classifications that are either correct or only one class away from the correct one).
Visualization of Workflows
Integrated AFM-ML Workflow for Biofilm Analysis
Biofilm Maturity Classification Logic
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials and Reagents for AFM-ML Biofilm Research
| Item | Function/Description | Example/Note |
|---|---|---|
| PFOTS-Treated Substrates | Creates a defined hydrophobic surface to study bacterial attachment dynamics [2]. | (Tridecafluoro-1,1,2,2-tetrahydrooctyl)trichlorosilane-treated glass coverslips. |
| Pantoea sp. YR343 | A model gram-negative, rod-shaped bacterium for studying flagella-mediated biofilm assembly [2]. | Isolated from the poplar rhizosphere; peritrichous flagella. |
| Staphylococcus aureus Strains | A model gram-positive bacterium for studying clinical biofilm-associated infections and maturity classification [26]. | Commonly used in studies of device-related infections. |
| Automated Large-Area AFM | Enables high-resolution imaging over millimeter-scale areas to capture biofilm heterogeneity [2]. | System with automated stage control and large-range piezoelectric actuators. |
| Image Stitching Software | Algorithms to seamlessly merge multiple contiguous AFM images into a single, large-area map [2]. | Often aided by machine learning for alignment with minimal matching features. |
| YOLO-based Object Detection Model | A deep learning architecture for real-time detection and localization of individual cells in images [27]. | YOLOv11 reported to achieve 93.8% mAP on blood cell detection tasks [27]. |
| Biofilm Maturity Classifier | A specialized deep learning algorithm for classifying biofilm development stages from AFM topographical data [26]. | Open-access desktop tool available; mean accuracy of 0.66 vs. ground truth [26]. |
| Formadicin | Formadicin, CAS:99150-60-4, MF:C30H34N4O16, MW:706.6 g/mol | Chemical Reagent |
| Digitolutein | Digitolutein, CAS:477-86-1, MF:C16H12O4, MW:268.26 g/mol | Chemical Reagent |
Optimizing Performance and Overcoming Challenges in HS-AFM Biofilm Imaging
Atomic Force Microscopy (AFM) has established itself as a powerful tool for structural biology, enabling the investigation of biological samples under physiologically relevant conditions. A paramount challenge, however, lies in minimizing sample disturbance to preserve the native state and viability of living cells during imaging. This is especially critical in the context of dynamic biofilm processes, where structural integrity and functional responses to environmental stimuli are the primary subjects of investigation. Excessive imaging forces can compress delicate structures, alter mechanical properties, and even disrupt vital processes, leading to artifactual data.
The evolution of AFM from a simple topographical imager to a multifunctional nanoscale laboratory has been driven by the need to overcome these challenges. Key developments, including dynamic imaging modes, high-speed AFM (HS-AFM), and functionalized cantilevers, have significantly reduced the invasiveness of measurements. This Application Note details the core strategies and provides detailed protocols for achieving low-invasiveness imaging, specifically tailored for research on dynamic biofilm formation and other live-cell interactions.
Core Principles of Low-Invasiveness AFM Operation
The perturbation exerted on a soft, living sample during AFM operation is governed by several interdependent factors. Minimizing disturbance requires a holistic approach that addresses each of these parameters.
Force Control and Cantilever Selection
The fundamental source of sample disturbance is the force applied by the AFM tip. Controlling this force requires careful selection and calibration of the cantilever.
- Low Spring Constants: For imaging living microbial cells and biofilms, cantilevers with small spring constants (0.01 - 0.10 N/m) are essential to limit the maximum force applied upon contact [28].
- Sharp, Clean Tips: A sharp tip (2–50 nm curvature radius) ensures high resolution while minimizing the contact area and, consequently, adhesive and friction forces [28].
- Advanced Feedback Modes: New operational modes, such as Bruker's "Scan Asyst," automatically and continuously adjust the applied force during scanning. This allows for high-resolution imaging of delicate samples like purple membranes and live cells for hours without damaging the sample [28].
Dynamic Imaging Modes
Operating in dynamic mode (also known as tapping or intermittent contact mode) is the cornerstone of low-invasiveness imaging for soft matter.
- Reduced Lateral Forces: In dynamic mode, the cantilever oscillates and only intermittently contacts the sample, dramatically reducing destructive lateral (shear) forces compared to static contact mode [4] [29]. This is critical for preventing the displacement of poorly adhered cells or the smearing of extracellular polymeric substances (EPS).
- Phase Imaging: This simultaneously acquired channel provides qualitative mapping of surface properties such as viscoelasticity and adhesion, offering insights into the sample's mechanical heterogeneity without additional invasive probing [4].
Table 1: Key Parameters for Low-Invasiveness AFM Imaging of Live Cells
| Parameter | Recommended Setting/Range | Rationale |
|---|---|---|
| Imaging Mode | Dynamic (Tapping) Mode | Minimizes lateral (shear) forces, preventing sample displacement and damage [4] [29]. |
| Cantilever Spring Constant | 0.01 - 0.10 N/m | Limits the maximum normal force exerted on soft, living samples [28]. |
| Setpoint Amplitude | As high as possible while maintaining stability | A lower setpoint increases tip-sample interaction time and force; use the minimum force needed for tracking. |
| Scan Rate | Optimized for sample tracking (typically 1-3 Hz) | Balances data acquisition speed with the feedback system's ability to accurately track topography. |
| Scanning Environment | Liquid (Appropriate Buffer) | Preserves native state, prevents dehydration, and reduces adhesion forces via screening [4] [28]. |
Advanced Methodologies for Minimal Disturbance
High-Speed AFM (HS-AFM) for Dynamic Processes
HS-AFM represents a breakthrough for directly observing biomolecular processes in real-time with sub-second temporal resolution.
- Principle: HS-AFM utilizes small, ultrafast cantilevers, faster scanners, and optimized feedback systems to achieve video-rate imaging (≥ 1 frame/second) [30] [29].
- Low Invasiveness: A key achievement of HS-AFM is that tip-sample interactions do not disturb biomolecules' functions. This has been demonstrated by visualizing myosin V walking on actin filaments and bacteriorhodopsin responding to light without functional impairment [30].
- Application to Biofilms: While challenging for large biofilm areas, HS-AFM is ideal for probing nanoscale dynamics at the interface of individual cells, such as the diffusion of membrane proteins (e.g., LacY) or the real-time assembly of EPS fibers [31].
Force-Distance Curve-Based AFM (FD-AFM)
This off-resonance mode separates topographical imaging from force measurement, offering a less invasive alternative to continuous contact mode.
- Methodology: At each pixel of the scan, the tip approaches, touches, and retracts from the surface, recording a force-distance curve. Topography is reconstructed from the contact points, while mechanical properties are mapped from the indentation and adhesion data [29].
- Benefits: By avoiding constant lateral dragging of the tip, FD-AFM significantly reduces shear stress. It allows for the simultaneous quantification of topography, adhesion, and viscoelasticity, providing a multiparametric view of the biofilm with minimal disturbance [32] [29].
Experimental Protocols for Live-Cell and Biofilm Imaging
Protocol 1: Immobilization of Microbial Cells for High-Resolution Imaging
Secure yet benign immobilization is the most critical step for successful live-cell AFM.
Method 1: Mechanical Entrapment in Porous Membranes
- Principle: Cells are physically trapped within a porous membrane with pore diameters similar to the cell size.
- Procedure:
- Place a microporous membrane (e.g., polycarbonate, pore size ~0.8-3 µm) on a solid support (e.g., glass slide).
- Apply a concentrated suspension of microbial cells onto the membrane.
- Gently filter the suspension, allowing cells to be captured in the pores.
- Rinse carefully with imaging buffer to remove non-immobilized cells.
- Transfer the membrane to the AFM liquid cell and submerge in buffer [4].
Method 2: Chemical Immobilization via Physiologically Benign Adhesives
- Principle: Uses natural adhesives or electrostatic interactions to anchor cells.
- Procedure:
- Prepare a coating solution of polydopamine or poly-L-lysine.
- Deposit a droplet onto a fresh mica or glass substrate for 10-30 minutes.
- Rinse thoroughly with deionized water to remove excess adhesive.
- Incubate a droplet of cell suspension on the coated substrate for 10-20 minutes.
- Rinse gently with imaging buffer to remove loosely attached cells [10]. Recent studies suggest that adding divalent cations (Mg²âº, Ca²âº) or glucose to the suspension can enhance attachment without compromising viability [4].
Protocol 2: Automated Large-Area AFM for Biofilm Heterogeneity
This protocol addresses the limitation of conventional AFM's small scan size, enabling the study of millimeter-scale biofilm organization without the invasiveness of stitching multiple manual scans.
- Principle: An AFM is integrated with a motorized stage and machine learning (ML) algorithms to automatically acquire and stitch hundreds of high-resolution images into a seamless millimeter-scale map [2].
- Procedure:
- Sample Preparation: Grow biofilms on a flat, adhesion-promoting substrate (e.g., PFOTS-treated glass) [2].
- Software Setup: Define the large area to be scanned. The ML algorithm optimizes the scan pattern with minimal overlap between individual images.
- Automated Acquisition: Initiate the automated run. The system sequentially images adjacent areas.
- Image Stitching and Analysis: ML-based algorithms seamlessly stitch the images. Subsequent analysis can automatically extract parameters like cell count, confluency, shape, and orientation over the entire large area [2].
Diagram 1: Workflow for automated large-area AFM imaging of biofilms.
Protocol 3: FluidFM for Biofilm-Surface Adhesion Measurements
This protocol uses FluidFM technology to measure adhesion forces between entire biofilms and surfaces, providing more realistic data than single-cell probes.
- Principle: A microfluidic cantilever with an aperture is used to aspirate and hold a biofilm-coated microbead. This probe is then used for force spectroscopy against a surface of interest [10].
- Procedure:
- Probe Fabrication: Grow a bacterial biofilm (e.g., Pseudomonas aeruginosa) on polystyrene microbeads.
- FluidFM Setup: Aspirate a biofilm-coated bead onto the aperture of a FluidFM cantilever by applying negative pressure.
- Force Spectroscopy: Approach the bead probe to the target surface (e.g., a modified filtration membrane) in liquid. Record multiple force-distance curves at different locations.
- Data Analysis: Analyze the retraction curves to quantify adhesion force, work of adhesion, and rupture events. Compare results to single-cell force spectroscopy (SCFS) to understand community-level adhesive behavior [10].
Table 2: The Scientist's Toolkit: Essential Reagents and Materials
| Item | Function/Application | Example/Specification |
|---|---|---|
| Polydopamine | A natural, non-denaturing adhesive for chemical immobilization of single cells or biofilms onto AFM substrates [10]. | Used to functionalize AFM tips for SCFS or to coat substrates for cell attachment. |
| Silicon Nitride Cantilevers | Standard probes for imaging in liquid. Low spring constants are critical for low-invasiveness. | Spring constant: 0.01 - 0.10 N/m; Tip radius: < 10 nm for high resolution [28]. |
| FluidFM Cantilever | Hollow cantilever for microfluidic manipulation. Enables biofilm-scale adhesion measurements via bead aspiration [10]. | Used with an external pressure controller to handle beads and single cells. |
| Microporous Membranes | For mechanical entrapment of cells, providing secure immobilization without chemical treatment [4]. | Polycarbonate membranes with pore sizes tailored to the microbe (e.g., 0.8 µm for bacteria). |
| PFOTS-Treated Glass | A hydrophobic surface treatment used to promote specific biofilm formation for structural studies [2]. | Creates a defined surface chemistry for studying early-stage biofilm assembly. |
The frontier of low-invasiveness AFM is moving towards even greater integration and automation. The application of machine learning and artificial intelligence is transforming AFM by optimizing scanning site selection, refining tip-sample interactions, and automating data analysis, thereby reducing human intervention and associated variability [2]. Furthermore, correlative microscopy combining HS-AFM or FD-AFM with super-resolution fluorescence techniques provides a comprehensive view, where dynamic structural data from AFM is overlaid with specific molecular localization from optics [29].
In conclusion, minimizing sample disturbance during AFM imaging of living cells is an achievable goal through a deliberate combination of appropriate hardware, operational modes, and sample preparation techniques. The strategies outlined here—from the foundational use of dynamic mode with soft cantilevers to the advanced application of HS-AFM, FD-AFM, and FluidFM—provide a robust toolkit for researchers. By adhering to these protocols, scientists can reliably investigate the dynamic architecture and functional mechanics of biofilms and other cellular systems in a state that closely mirrors their native, physiological condition.
The investigation of dynamic biofilm processes using high-speed Atomic Force Microscopy (HS-AFM) necessitates precise control over the experimental environment. Biofilms are complex, three-dimensional microbial communities that exhibit spatial and temporal heterogeneity in their structure, composition, and function [2] [33]. Their development and response to external stimuli are profoundly influenced by environmental conditions such as temperature and the chemical composition of the surrounding liquid medium [1]. Uncontrolled variations in these parameters can introduce significant artifacts, alter biofilm physiology, and compromise the validity of nanoscale measurements. Therefore, robust protocols for maintaining thermal stability and executing buffer exchange are critical for obtaining reproducible, high-resolution data that accurately reflects biofilm dynamics. This document provides detailed application notes and protocols for managing these variables within the broader context of a thesis on HS-AFM imaging of dynamic biofilm processes.
The Critical Role of Environmental Control in HS-AFM Biofilm Imaging
The integrity of HS-AFM data is inextricably linked to the stability of the sample environment. For biofilms, which are highly responsive to their surroundings, this is particularly crucial. Temperature fluctuations can directly affect bacterial metabolism, growth rates, and the physical properties of the extracellular polymeric substance (EPS) [33]. Even minor deviations can lead to changes in biofilm stiffness, adhesion, and overall architecture, which are key parameters measured by AFM. Furthermore, the process of introducing chemical stimuli, nutrients, or antibiotics during imaging requires solution exchange protocols that minimize mechanical disturbance to the delicate biofilm structure. Advances in AFM, such as the use of Quantitative Imaging (QI) mode in liquid, have enabled the visualization of native bacterial structures like nanotubes and flagella without aggressive immobilization [34]. These techniques, however, depend entirely on a stable substrate and controlled liquid cell conditions to achieve high-quality, high-resolution imaging of living cells in physiological conditions.
Table 1: Key Nanomechanical and Imaging Parameters for Biofilm AFM in Liquid
| Parameter | Typical Value / Range | Measurement Technique | Biological Significance |
|---|---|---|---|
| Young's Modulus (Bacterium) | ~0.236 ± 0.05 N/m [34] | Force curve analysis via Hertz/Sneddon model | Indicator of cellular stiffness and structural integrity. |
| Young's Modulus (Nanotubes) | Lower than main cell body [34] | Force curve analysis via Hertz/Sneddon model | Suggests flexibility, essential for intercellular communication. |
| AFM Tip Speed (QI Mode) | 125 µm/s [34] | Quantitative Imaging mode setting | Balances imaging speed with force control to prevent sample damage. |
| Indentation Speed | 17–175 mN/s [34] | Force spectroscopy setting | Controls the rate of loading during nanomechanical probing. |
| Bacterial Cell Dimensions (Pantoea sp.) | ~2 µm (length) x ~1 µm (diameter) [2] | High-resolution topographical imaging | Provides baseline morphological data for the studied organism. |
| Flagella Height | ~20–50 nm [2] | High-resolution topographical imaging | Visualizes key appendages for attachment and motility. |
Table 2: Experimental Conditions for Environmental Control
| Parameter | Optimal Setting | Protocol / Equipment | Impact on Experiment |
|---|---|---|---|
| Temperature Control | 24.0 ± 0.2 °C [34] | Thermostatted ECCell or liquid cell | Maintains bacterial metabolic activity and prevents thermal drift. |
| Substrate for Adhesion | Indium-Tin-Oxide (ITO) coated glass [34] | Non-perturbative sample preparation | Provides smooth, hydrophobic surface for stable cell adhesion without chemical fixation. |
| Cantilever Stiffness | 0.3 N/m [34] | PPP-CONTPt probes (Nanosensors) | Soft enough for reliable force spectroscopy on soft biological samples. |
| Imaging Mode | Quantitative Imaging (QI) | Fast approach/retract mode in liquid | Enables concurrent topographical and nanomechanical mapping with high resolution. |
Experimental Protocols
Protocol 1: Temperature-Stabilized HS-AFM Imaging of Living Biofilms
This protocol is designed for nanomechanical mapping of living biofilms, such as Rhodococcus wratislaviensis, under physiological conditions with minimal perturbation [34].
Key Research Reagent Solutions:
- Bacterial Strain: Rhodococcus wratislaviensis (or other relevant biofilm-forming strain).
- Growth Medium: Suitable liquid culture medium (e.g., Lysogeny Broth).
- Imaging Buffer: Fresh culture medium or a physiologically compatible buffer (e.g., PBS).
- Substrate: Indium-Tin-Oxide (ITO)-coated glass slides. ITO provides a smooth, hydrophobic surface that promotes bacterial adhesion without requiring chemical fixation.
- AFM Probes: Soft cantilevers (e.g., PPP-CONTPt, Nanosensors) with a nominal spring constant of 0.3 N/m.
Methodology:
- Sample Preparation: Pipette 500 µL of bacterial culture during its exponential growth phase directly onto an ITO-coated glass substrate mounted in an ElectroChemical Cell (ECCell).
- Temperature Equilibration: Place the ECCell into the AFM instrument and allow the system to equilibrate to the target temperature (e.g., 24.0 ± 0.2 °C) for at least 15-20 minutes before engaging the probe.
- AFM Imaging and Data Acquisition:
- Engage the AFM probe in the liquid medium using a fast-speed approach/retract mode, such as Quantitative Imaging (QI) mode.
- Set the imaging parameters: total extension of 600 nm at a constant speed of 125 µm/s, with an indentation speed between 17–175 mN/s.
- Acquire height images and concurrent mechanical property maps (Young's modulus) by performing force curves at each pixel.
Protocol 2: Buffer Exchange for In-Situ Stimulation During HS-AFM
This protocol outlines a procedure for introducing chemical stimuli or altering the buffer composition during time-lapse HS-AFM imaging to monitor dynamic biofilm responses.
Key Research Reagent Solutions:
- Initial Imaging Buffer: Standard buffer or growth medium.
- Stimulus Buffer: Initial imaging buffer containing the compound of interest (e.g., antibiotic, surfactant, nutrient).
- Wash Buffer: Initial imaging buffer without additives.
Methodology:
- Baseline Imaging: Establish stable HS-AFM imaging of the biofilm region of interest in the initial imaging buffer, following Protocol 1.
- System Preparation: Connect a syringe or peristaltic pump containing the stimulus buffer to the inlet port of the AFM liquid cell. Connect a waste line to the outlet port, ensuring it leads to a waste container.
- Controlled Exchange:
- Pause the scanning process temporarily to minimize tip disturbance during fluid flow.
- Initiate a slow, continuous flow (e.g., 0.5 - 1 mL/min) of the stimulus buffer into the liquid cell, allowing the old buffer to be displaced through the outlet. A total volume exchange of 5-10 times the liquid cell volume is recommended to ensure complete replacement.
- Resume HS-AFM imaging immediately after the exchange is complete to capture the biofilm's real-time response.
- Post-Stimulus Wash (Optional): To study recovery or remove the stimulus, repeat step 3 using the wash buffer.
Workflow and System Visualization
HS-AFM Biofilm Stimulation Workflow
Nanoscale Biofilm Components and AFM Interaction
Atomic force microscopy (AFM) has become an indispensable tool for probing the dynamic processes of biofilm formation, structure, and response to environmental cues at the nanoscale. Its capability to operate under physiological liquid conditions makes it particularly valuable for studying biofilms in their native hydrated state. However, researchers face significant technical challenges when performing high-speed AFM imaging in liquids, where artifacts such as thermal drift, electronic noise, and hydrodynamic drag forces can compromise data quality and biological interpretation. These artifacts are particularly problematic when investigating time-sensitive biofilm processes, including initial bacterial attachment, microcolony development, and extracellular polymeric substance (EPS) secretion. This application note provides detailed protocols and analytical frameworks for identifying, minimizing, and correcting these common artifacts, enabling more reliable nanoscale investigation of dynamic biofilm systems for therapeutic development.
Artifact Fundamentals and Impact on Biofilm Research
Characterizing Common AFM Artifacts
Understanding the origin and manifestation of each artifact type is crucial for developing effective mitigation strategies. The following table summarizes the primary artifacts affecting AFM imaging in liquid environments.
Table 1: Common AFM Artifacts in Liquid Environments and Their Characteristics
| Artifact Type | Primary Causes | Visual Manifestation | Impact on Biofilm Imaging |
|---|---|---|---|
| Thermal Drift | Temperature fluctuations, scanner creep, uneven thermal expansion | Image stretching, compression, distortion; blurred features over time | Misrepresentation of bacterial cell dimensions, inaccurate tracking of dynamic processes |
| Electronic Noise | Electromagnetic interference, ground loops, detector noise | Horizontal striations, random pixel variations, high-frequency image components | Obscures fine structural details (flagella, pili, EPS fibers); reduces signal-to-noise ratio |
| Hydrodynamic Drag | Viscous resistance between cantilever and fluid; higher in liquids than air | Force curve offsets, baseline drift, erroneous adhesion/mechanical measurements | Distorts nanomechanical property mapping; interferes with peak force detection in soft biofilm matrix |
Experimental Workflow for Artifact Mitigation
The following diagram outlines a systematic workflow for addressing these artifacts throughout an AFM experiment, from preparation through to final image processing.
Protocols for Artifact Identification and Correction
Thermal Drift Management Protocol
Thermal drift causes progressive distortion in AFM images as dimensional stability is compromised by temperature fluctuations during scanning. This is particularly problematic for time-lapse studies of biofilm development where accurate dimensional tracking is essential.
Experimental Protocol: Drift Quantification and Correction
- System Stabilization: Allow the AFM system to equilibrate for at least 60 minutes in the imaging environment before beginning experiments. Ensure temperature stability of ±0.1°C in the laboratory.
- Drift Rate Calculation: Image a standard calibration grating with well-defined feature spacing at the beginning and end of imaging sessions. Calculate drift rates from feature displacement using the formula: Drift Rate = (Δx² + Δy²)¹áŸÂ² / Δt where Δx and Δy are displacement components and Δt is the time interval between measurements.
- Scanner Conditioning: Perform slow, full-range scans of the piezoelectric scanner for 5-10 minutes before high-resolution imaging to minimize creep effects.
- Post-Acquisition Correction: Apply affine transformation algorithms in analysis software (e.g., Gwyddion) using known feature dimensions as reference points.
Electronic Noise Reduction Protocol
Electronic noise introduces high-frequency interference that can obscure critical nanoscale features of bacterial cells and biofilm matrix components during imaging.
Experimental Protocol: Noise Identification and Suppression
- Grounding Verification: Ensure all system components share a common ground point. Use a dedicated grounding rod for sensitive AFM measurements.
- Environmental Shielding: Enclose the AFM system in a Faraday cage to block electromagnetic interference. Use shielded cables for all electrical connections.
- Detection Optimization: Adjust laser alignment and photodetector positions to maximize sum and deflection signals while minimizing differences.
- Scan Parameter Optimization: Set scan rates below the system's resonant frequency in liquid (typically 0.5-1.5 Hz for high-resolution imaging). Adjust integral and proportional gains to maintain stability without introducing oscillation.
- Advanced Processing: Apply convolutional neural networks (CNN) trained on synthetic AFM data to suppress noise while preserving surface structures [35]. The network architecture should utilize two scanning directions with complementary errors as input for improved reconstruction.
Hydrodynamic Drag Compensation Protocol
Hydrodynamic drag forces in liquid environments can significantly distort force measurements and interfere with accurate peak force detection, critically affecting nanomechanical characterization of biofilms.
Experimental Protocol: Drag Force Minimization and Correction
- Cantilever Selection: Choose triangular cantilevers rather than rectangular designs, as they generate significantly lower drag forces [36]. Select shorter cantilevers (100 μm) with lower spring constants (0.06-0.32 N/m) for reduced flow resistance.
- Liquid Medium Optimization: Use ultrapure water instead of ethanol-water mixtures when possible, as drag forces increase with ethanol concentration [36].
- Imaging Parameter Adjustment: Reduce oscillation amplitudes (50-150 nm) and frequencies (0.8-1.2 kHz) to lower maximum tip velocities, thereby minimizing drag forces that scale linearly with velocity [36].
- Force Baseline Correction: Characterize the force-distance curve baseline away from the sample before engaging. Subtract the hydrodynamic contribution from measured force curves using the relationship: F_drag = k·v where k is the drag coefficient and v is the tip velocity.
- Temperature Control: Maintain constant temperature during imaging, as viscosity changes with temperature directly affect drag forces.
Table 2: Hydrodynamic Drag Forces for Different Cantilever Geometries in Ultrapure Water
| Cantilever Shape | Length (μm) | Spring Constant (N/m) | Relative Drag Force | Recommended Applications |
|---|---|---|---|---|
| Triangular | 100 | 0.32 | 1.0× | High-resolution cell imaging |
| Triangular | 200 | 0.08 | 1.8× | Soft biofilm matrix mapping |
| Rectangular | 100 | 0.48 | 2.5× | Stiffness measurements |
| Rectangular | 200 | 0.06 | 3.2× | Low-force adhesion studies |
Advanced Integration: Machine Learning Approaches
Recent advances in machine learning provide powerful new approaches for artifact correction that complement traditional methods. Convolutional neural networks (CNNs) can be trained to recognize and remove artifacts while preserving legitimate sample features.
Experimental Protocol: CNN Implementation for Artifact Correction
- Network Architecture: Implement a ResU-Net architecture, which combines residual connections with U-Net skip connections, ideal for AFM image processing [35].
- Synthetic Data Generation: Create training datasets by generating random topographies in the spectral domain followed by inverse Fourier transform. Artificially introduce common AFM artifacts including noise, drift distortions, and tip effects.
- Model Training: Train the network using pairs of artifact-free synthetic topographies and corresponding images with simulated artifacts. Use physical AFM samples scanned at multiple rotations for validation.
- Application: Process acquired AFM images by inputting both forward and backward scan directions. The network outputs reconstructed images with suppressed artifacts while preserving critical surface features such as edges and fine structures.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Research Reagent Solutions for AFM Biofilm Studies
| Item | Function/Application | Example Specifications |
|---|---|---|
| PFOTS-treated Glass | Hydrophobic surface for controlled bacterial attachment | (3,3,3-Trifluoropropyl)trichlorosilane treated coverslips |
| Silicon Nitride Probes | Bioimaging in liquid environments | Triangular cantilevers, spring constant: 0.08-0.32 N/m |
| Polydimethylsiloxane (PDMS) Stamps | Cell immobilization for aqueous imaging | Microstructured stamps (1.5-6 µm wide, 1-4 µm deep) |
| Functionalization Reagents | Surface modification for specific adhesion studies | Poly-L-lysine, trimethoxysilyl-propyl-diethylenetriamine |
| Imaging Buffer Solutions | Maintain physiological conditions during imaging | Phosphate buffered saline with divalent cations (Mg²âº, Ca²âº) |
Effective management of AFM artifacts in liquid environments is essential for obtaining reliable, high-resolution data on dynamic biofilm processes. By implementing the systematic protocols outlined in this application note—including thermal stabilization procedures, electronic noise suppression techniques, hydrodynamic drag compensation methods, and machine learning approaches—researchers can significantly improve data quality and biological relevance. These advanced artifact mitigation strategies enable more accurate investigation of fundamental biofilm properties, from initial bacterial adhesion and structural organization to mechanical response and antimicrobial susceptibility, ultimately accelerating therapeutic development against biofilm-associated infections.
In the context of high-speed atomic force microscopy (AFM) imaging of dynamic biofilm processes, the integrity of the data is fundamentally dependent on the probe's condition and correct selection. Biofilms, as complex microbial communities encased in extracellular polymeric substances (EPS), present a challenging landscape for nanoscale investigation [2] [37]. The inherent heterogeneity and delicate, dynamic nature of biofilms demand imaging techniques that are not only high-resolution but also minimally disruptive [3]. While AFM provides critically important high-resolution insights on structural and functional properties at the cellular and even sub-cellular level, its effectiveness is heavily influenced by the probe [2]. The probe is the primary point of interaction between the instrument and the biological sample; its geometry directly dictates resolution, and its surface chemistry influences tip-sample interactions. Furthermore, probe conditioning—the process of preparing and maintaining a stable, sharp tip—is not merely a preliminary step but a continuous requirement for acquiring consistent, high-fidelity data over the extended timescales necessary to capture biofilm dynamics [2]. This document outlines detailed protocols and evidence-based guidelines for probe selection and conditioning, specifically tailored for researchers investigating the assembly and behavior of biofilms.
The Role of AFM in Biofilm Research
Atomic force microscopy (AFM) enables the imaging of cell structures, interactions, and mechanical properties at the nanoscale without extensive sample preparation, and can be used under physiological conditions [2] [3]. By scanning a sharp probe over the surface and measuring the forces between the probe and the sample, AFM provides nanometer-scale topographical images as well as quantitative maps of nanomechanical properties [2]. This allows AFM to reveal structural features of biofilms that surpass the resolution of optical or electron beam-based microscopy, such as membrane protrusions, surface proteins, and the fine structures of the EPS that form the biofilm matrix [2] [37].
However, conventional AFM has limitations for biofilm studies. Its small imaging area (typically less than 100 µm), restricted by piezoelectric actuator constraints, limits its capacity to study large, millimeter-scale biofilm structures [2]. This scale mismatch makes it difficult to capture the full spatial complexity of biofilms. Furthermore, the slow scanning process and labor-intensive operation hinder the capture of dynamic structural changes [2]. Emerging approaches, such as automated large-area AFM aided by machine learning (ML), are beginning to address these limitations by enabling high-resolution imaging over millimeter-scale areas and automating routine tasks, thus allowing for continuous, multi-day experiments [2] [1]. Despite these advancements, the fundamental importance of the probe remains unchanged.
Key AFM Imaging Modes for Biofilm Studies
The following table summarizes the primary AFM modes used in biofilm research.
Table 1: Key AFM Imaging Modes for Biofilm Studies
| Imaging Mode | Principle | Key Applications in Biofilm Research | Advantages for Biofilms |
|---|---|---|---|
| Contact Mode | The tip scans in constant physical contact with the sample. | Topographical imaging of robust features; surface roughness mapping of conditioning films on plastics [38]. | Simple operation; high scan speeds suitable for some dynamic processes. |
| Tapping Mode | The cantilever oscillates at resonance, briefly tapping the surface. | High-resolution imaging of delicate samples; visualizing individual bacterial cells, flagella, and pili [2]. | Minimizes lateral forces, reducing sample damage; ideal for soft, adhesive biofilm components. |
| Force Spectroscopy | The tip approaches, contacts, and retracts from the surface while measuring force. | Quantifying adhesion forces between cells and surfaces; mapping local mechanical properties (e.g., stiffness, elasticity) of the EPS matrix [3] [37]. | Provides quantitative, nanoscale mechanical data; can be used under physiological conditions. |
Probe Selection for Biofilm Imaging
The selection of an appropriate AFM probe is a critical first step that dictates the quality and type of data that can be acquired. The choice must balance the need for high resolution with the soft, adhesive, and often heterogeneous nature of biofilms.
Probe Characteristics and Their Impact on Imaging
The following table details the key characteristics to consider when selecting a probe for biofilm studies.
Table 2: AFM Probe Selection Guide for Biofilm Imaging
| Probe Characteristic | Impact on Data Acquisition | Recommended Specification for Biofilms |
|---|---|---|
| Nominal Tip Radius | Determines lateral resolution. A sharper tip resolves finer details. | < 10 nm for high-resolution imaging of cellular features and appendages like flagella (~20-50 nm in height) [2]. |
| Cantilever Stiffness (k) | Influences force control and sample damage. Softer levers reduce indentation on soft samples. | 0.1 - 0.5 N/m for imaging in liquid [3]. Stiffer levers (∼0.06 N/m) may be used for contact mode in liquid [38]. |
| Resonant Frequency (in liquid) | Critical for tapping mode performance. Higher frequencies can enable faster scanning. | 10 - 50 kHz for optimal performance in fluid environments [37]. |
| Cantilever Material | Affects reflectivity and chemical compatibility. | Silicon Nitride (SiN) is common for its favorable properties in biological liquids [38]. |
| Coating | Can enhance reflectivity or enable functionalized imaging. | Uncoated or reflective coating (e.g., gold) for standard imaging. |
The Scientist's Toolkit: Essential Reagents and Materials
Table 3: Research Reagent Solutions for AFM Probe Conditioning and Biofilm Imaging
| Item | Function/Application in Protocol |
|---|---|
| Silicon Nitride (SiN) Probes | Standard probe for imaging in physiological liquids; biocompatible and with appropriate spring constants for soft samples [38]. |
| UV-Ozone Cleaner | Equipment for dry cleaning of probes to remove organic contaminants via oxidative processes. |
| Piranha Solution | CAUTION: Highly reactive and hazardous. A mixture of sulfuric acid (Hâ‚‚SOâ‚„) and hydrogen peroxide (Hâ‚‚Oâ‚‚) used for intensive cleaning of metal-coated probes to remove all organic residues. |
| Ethanol (≥99%) | Solvent for rinsing probes after cleaning or exposure to biological samples. |
| Deionized Water | Solvent for final rinsing steps to remove salts and other residues. |
| Plasma Cleaner (Oxygen plasma) | Equipment for a highly effective and gentle final cleaning step, rendering the probe hydrophilic and pristine. |
| Calibration Grating | A sample with known topography (e.g., TGQ1, TGXYZ) used for validating tip sharpness and scanner calibration. |
Experimental Protocol for Probe Conditioning and Validation
This protocol provides a detailed, step-by-step methodology for conditioning AFM probes to ensure consistent, high-resolution data acquisition during biofilm imaging.
Protocol: Probe Conditioning and Performance Validation
Objective: To clean, characterize, and validate an AFM probe for high-resolution imaging of biofilms, minimizing artifacts and ensuring data consistency.
Materials and Equipment:
- New or used AFM probe (e.g., SiN, nominal frequency ~30 kHz in liquid, stiffness ~0.1 N/m).
- UV-Ozone cleaner or Plasma cleaner.
- ACS-grade ethanol and deionized water.
- Compressed air or nitrogen gas duster.
- Standard calibration grating (e.g., silicon with periodic structures like trenches or dots).
- AFM instrument.
Procedure:
Part A: Initial Visual Inspection and Cleaning
- Visual Inspection: Under an optical microscope at high magnification (50x-100x), inspect the probe and cantilever for large contaminants, cracks, or chips. Discard the probe if any damage is visible.
- Dry Cleaning: a. Use a stream of clean, compressed air or nitrogen directed at the probe chip at a shallow angle to dislodge loose particulate matter. b. Place the probe in a UV-Ozone cleaner for 10-15 minutes. This will remove thin organic contaminant layers through photo-oxidation.
- Wet Cleaning (if necessary): a. If the probe is heavily contaminated (e.g., with dried salt or polymer from a previous biofilm scan), perform a solvent clean. b. CAUTION: This can damage certain coatings. For uncoated SiN probes, immerse the probe in ethanol for 5 minutes, followed by immersion in deionized water for 5 minutes. c. Dry the probe completely using a gentle stream of clean, compressed air or nitrogen.
Part B: In-Situ Conditioning and Integrity Check
- Mount the Probe: Carefully mount the cleaned probe in the AFM holder according to the manufacturer's instructions.
- Laser Alignment: Align the laser spot on the end of the cantilever and maximize the sum signal. Adjust the photodetector to center the deflection signal.
- Thermal Tune: Engage the cantilever in air (or the intended imaging medium) and perform a thermal tune to determine its actual resonant frequency and quality factor (Q-factor). Compare these values to the manufacturer's specifications. A significant deviation may indicate contamination or damage.
- Engage on a Clean, Hard Surface: a. Use a clean silicon wafer or mica surface. b. Engage the probe and capture a small-scale (1x1 µm) image in tapping mode. c. Assess the image for "double tipping" or other artifacts that suggest a contaminated or broken tip.
Part C: Tip Sharpness Validation Using a Calibration Grating
- Image the Grating: Image a sharp-featured calibration grating (e.g., a silicon grating with sharp spikes or sharp trenches) using the same parameters intended for biofilm imaging.
- Analyze the Image: a. Measure the tip's contribution to the image by analyzing the aspect ratio of the features. A sharp, clean tip will produce images with near-vertical sidewalls and sharp peaks. b. A dull or contaminated tip will produce images with flattened peaks and widened, sloped trenches, a phenomenon known as tip-broadening.
- Quantify Tip Geometry: Use the AFM software's tip reconstruction algorithm (if available) to generate a 3D model of the tip's apex from the grating image. A tip radius of less than 10 nm is desirable for high-resolution biofilm work [2].
Part D: Final Performance Check and Readiness
- Functional Test: Image a test sample of known, fine structure relevant to biofilms, such as isolated bacterial cells or a protein-coated surface.
- Assess Resolution: Verify that the probe can resolve expected features, such as the height of bacterial cells (~1-2 µm) and the fine structure of flagella (expected height ~20-50 nm) [2].
- The probe is now conditioned and validated for high-resolution biofilm imaging.
Workflow for Probe Management in High-Speed AFM Studies
The following diagram illustrates the logical workflow for probe selection, conditioning, and data acquisition, integrating machine learning to enhance efficiency as referenced in recent literature [2] [1].
Probe Management and Conditioning Workflow
The path to consistent, high-resolution data in high-speed AFM imaging of dynamic biofilms is paved with meticulous attention to the probe. As this application note has detailed, a systematic approach encompassing informed probe selection, rigorous conditioning protocols, and continuous performance validation is non-negotiable. The integration of machine learning for automated probe conditioning represents the cutting edge, promising to mitigate drift and reduce human intervention during long-term experiments [2] [1]. By adhering to these guidelines, researchers can ensure that their AFM data accurately reflects the complex and dynamic world of biofilms, thereby accelerating discoveries in drug development, environmental science, and microbial ecology.
Validating Findings and Comparative Analysis: HS-AFM in the Broader Microscopy Landscape
Within the broader research on high-speed atomic force microscopy (HS-AFM) of dynamic biofilm processes, benchmarking against established imaging techniques is a critical step. This application note details the methodologies for a comparative analysis of Scanning Electron Microscopy (SEM), Confocal Laser Scanning Microscopy (CLSM), and Raman Spectroscopy, framing them within the context of validating and complementing HS-AFM data. Biofilms, as complex microbial communities encased in an extracellular polymeric substance (EPS), exhibit dynamic spatial and chemical heterogeneities that no single technique can fully characterize [2] [39]. This document provides detailed protocols and quantitative comparisons to guide researchers in designing multi-modal imaging strategies that leverage the strengths of each technology, thereby accelerating drug development against biofilm-associated infections.
The following table summarizes the core capabilities and limitations of each technique, providing a benchmark for their application in biofilm research.
Table 1: Benchmarking Established Techniques for Biofilm Analysis
| Technique | Best For / Key Application | Resolution (approx.) | Key Advantages | Inherent Limitations |
|---|---|---|---|---|
| Scanning Electron Microscopy (SEM) | High-resolution surface topography and ultrastructure [40]. | 1-10 nm [40] | Unparalleled image quality, high magnification, and great depth of field for surface details [40]. | Requires extensive sample preparation (dehydration, fixation, coating) risking artifacts like EPS collapse [40]. |
| Confocal Laser Scanning Microscopy (CLSM) | 3-D architecture, live/dead cell distribution, and real-time, in-situ monitoring of hydrated biofilms [39] [40]. | ~200-300 nm (lateral) [40] | Non-destructive optical sectioning for 3D reconstruction; ability to use fluorescent probes for functional imaging [40]. | Limited by photobleaching; fluorescence can be interfered with by intrinsic biofilm signals [40]. |
| Raman Spectroscopy | Molecular-level chemical analysis of biofilm composition (e.g., EPS proteins, lipids, nucleic acids) without staining [39]. | Diffraction-limited (~1 µm) | Provides label-free, chemical fingerprinting of biofilm constituents; can be combined with microscopy for spatial mapping [39]. | Relatively weak signal; can be susceptible to fluorescence interference; may require high laser power risking photodamage [39]. |
| Atomic Force Microscopy (AFM) | Nanoscale surface topography and quantitative nanomechanical properties (e.g., stiffness, adhesion) under physiological conditions [2] [4]. | Sub-nanometer (vertical) [4] | Operates in liquids on native, unfixed samples; measures mechanical properties and interaction forces [4]. | Small imaging area can be unrepresentative; slow scan speed for conventional AFM limits dynamic studies [2]. |
The following workflow illustrates a proposed integrated approach for a comprehensive biofilm analysis, leveraging the strengths of each technique.
Diagram 1: Workflow for multi-modal biofilm analysis.
Experimental Protocols
Protocol for SEM Analysis of Biofilm Ultrastructure
This protocol is designed for high-resolution visualization of biofilm surface morphology and cellular arrangement.
1. Sample Preparation:
- Fixation: Immerse biofilm-covered substrate in a solution of 2.5% glutaraldehyde in a 0.1 M sodium cacodylate buffer (pH 7.4) for a minimum of 2 hours at 4°C. For enhanced EPS preservation, include 1% ruthenium red or tannic acid in the fixative [40].
- Washing: Rinse the sample three times (10 minutes each) with the same buffer to remove excess fixative.
- Dehydration: Subject the sample to a graded ethanol series (30%, 50%, 70%, 80%, 90%, 95%, and three times 100%), allowing 10-15 minutes per step.
- Drying: Critical Point Dry (CPD) the sample using liquid COâ‚‚ as the transition fluid to avoid surface tension artifacts. Alternatively, use hexamethyldisilazane (HMDS) for a faster, air-drying protocol [40].
- Mounting and Coating: Mount the sample on an aluminum stub using conductive carbon tape. Sputter-coat with a 10-20 nm layer of gold/palladium to render the sample conductive.
2. Data Acquisition:
- Load the sample into a conventional or field-emission SEM.
- Set the accelerating voltage to 5-15 kV for an optimal balance between resolution and minimizing sample charging.
- Acquire images at various magnifications (e.g., 1,000x to 50,000x) to capture both overall biofilm architecture and fine ultrastructural details.
Protocol for CLSM for 3D Biofilm Architecture and Viability
This protocol enables non-destructive, three-dimensional imaging of biofilm structure and cell viability under hydrated conditions.
1. Sample Preparation and Staining:
- Grow biofilms on surfaces compatible with microscopy (e.g., glass-bottom dishes).
- Prepare a staining solution containing SYTO 9 (labels all bacterial nucleic acids) and propidium iodide (PI) (labels only dead/damaged cells) as per the manufacturer's instructions for the LIVE/DEAD BacLight kit.
- Gently replace the culture medium in the biofilm dish with the staining solution.
- Incubate in the dark for 15-20 minutes at room temperature.
2. Data Acquisition:
- Use a confocal laser scanning microscope equipped with appropriate laser lines (e.g., 488 nm) and emission filters (e.g., 500-550 nm for SYTO 9, 600-650 nm for PI).
- Set the objective lens to a suitable magnification (e.g., 40x or 63x water-immersion objective).
- Define the Z-stack by setting the upper and lower limits of the biofilm. Set the step size to 1-2 µm to ensure adequate resolution in the Z-dimension.
- Acquire the Z-stack series. Most CLSM software will allow for the simultaneous collection of both fluorescence channels.
3. Data Analysis:
- Use image analysis software (e.g., ImageJ, IMARIS, or COMSTAT) to reconstruct the 3D volume and calculate quantitative parameters such as:
- Biovolume: Total volume of the biofilm (µm³/µm²).
- Thickness: Maximum and average thickness of the biofilm (µm).
- Surface Area to Biovolume Ratio: Indicator of biofilm complexity.
- Viability Ratio: Ratio of live (green) to dead (red) signal intensity.
Protocol for Raman Spectroscopy for Chemical Mapping
This protocol outlines the steps for acquiring chemical fingerprints from a biofilm to determine its molecular composition.
1. Sample Preparation:
- Grow biofilms on an optically flat, non-fluorescent substrate such as calcium fluoride (CaFâ‚‚) or gold-coated slides.
- For Surface-Enhanced Raman Spectroscopy (SERS), decorate the substrate with plasmonically active nanoparticles (e.g., silver or gold colloids) prior to biofilm growth to significantly enhance the Raman signal [41].
- Rinse the sample gently with ultrapure water to remove loosely attached planktonic cells and media components. Air-dry or analyze under hydrated conditions as required.
2. Data Acquisition:
- Place the sample on the microscope stage of the Raman spectrometer.
- Focus the laser onto the biofilm surface using a high-magnification objective (e.g., 50x or 100x).
- Set acquisition parameters:
- Laser Wavelength: 532 nm or 785 nm are common choices; 785 nm reduces fluorescence.
- Laser Power: Keep as low as possible (e.g., 1-10 mW at the sample) to avoid photodamage.
- Grating: Choose an appropriate grating for the desired spectral resolution.
- Integration Time: Typically 1-10 seconds per spectrum.
- To create a chemical map, define a region of interest and a step size (e.g., 1 µm). The instrument will automatically acquire a full spectrum at each point.
3. Data Analysis:
- Pre-process spectra: subtract fluorescence background (e.g., using a polynomial fit), correct for cosmic rays, and normalize.
- Identify characteristic Raman bands for key biofilm components:
- Polysaccharides: ~480, 850, 940, 1100 cmâ»Â¹
- Proteins: Amide I (~1650 cmâ»Â¹), Amide III (~1240-1300 cmâ»Â¹), phenylalanine (~1000 cmâ»Â¹)
- Nucleic Acids: ~785 cmâ»Â¹ (DNA backbone)
- Use multivariate analysis (e.g., Principal Component Analysis - PCA) or cluster analysis (e.g., K-means) to classify spectra and generate false-color maps showing the spatial distribution of chemical components.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 2: Key Research Reagents and Materials for Biofilm Imaging
| Item Name | Function / Application | Brief Rationale |
|---|---|---|
| PFOTS-treated Glass | Surface for biofilm growth in AFM/SEM studies [2]. | Creates a defined hydrophobic surface to study early bacterial attachment and biofilm assembly dynamics [2]. |
| Ruthenium Red (RR) | Additive for SEM sample fixation [40]. | Binds to and helps preserve acidic polysaccharides in the EPS matrix during preparation, reducing structural collapse [40]. |
| LIVE/DEAD BacLight Viability Kit | Fluorescent staining for CLSM [40]. | Contains SYTO 9 and propidium iodide (PI) to differentially label live (intact membrane) and dead (compromised membrane) cells in a biofilm. |
| Gold Nanoparticles (e.g., 60 nm) | Substrate for Surface-Enhanced Raman Spectroscopy (SERS) [41]. | Provides plasmonic enhancement of the inherently weak Raman signal, enabling detection of trace metabolites and molecules within the biofilm. |
| Polydimethylsiloxane (PDMS) Stamps | Micro-patterned substrates for AFM/CLSM [4]. | Enables precise mechanical immobilization of microbial cells for stable, high-resolution imaging under fluid conditions. |
| Glutaraldehyde (2.5% in Buffer) | Primary fixative for SEM and TEM [40]. | Cross-links proteins and other biomolecules, stabilizing the 3D structure of the biofilm against dehydration and vacuum. |
Integrated Data Interpretation and Correlation with HS-AFM
The synergy between these techniques provides a powerful framework for interpreting HS-AFM data on dynamic biofilm processes. CLSM can first identify regions of interest based on biofilm thickness and viability. Raman spectroscopy can then be applied to these regions to determine if variations in structure correlate with changes in chemical composition, such as EPS protein-to-polysaccharide ratios. SEM can provide ultra-high-resolution snapshots of the surface morphology in these distinct regions.
Subsequently, HS-AFM can be targeted to these pre-characterized areas to capture real-time, nanoscale dynamics. For instance, the mechanical properties (e.g., elasticity measured by HS-AFM nanoindentation) of a region identified by Raman as EPS-rich can be directly correlated with its chemical identity. This multi-modal approach transforms HS-AFM from a tool that provides exquisite temporal and spatial resolution of topography and mechanics into one whose findings are grounded in comprehensive structural and chemical context. This is crucial for drug development, as it allows for the direct visualization of how anti-biofilm compounds alter not just the physical structure and mechanics of a biofilm, but also its chemical heterogeneity and cellular viability.
The study of dynamic biofilm processes, such as initial bacterial attachment, community development, and response to antimicrobial agents, requires observation across multiple spatial and temporal scales. High-speed atomic force microscopy (HS-AFM) provides unparalleled nanoscale topographical and mechanical data of bacterial cell surfaces, enabling researchers to visualize processes like the role of flagella in early-stage biofilm formation and the emergence of complex cellular arrangements [2]. However, a significant limitation of AFM is its inherent inability to visualize internal cellular events and biochemical signaling that accompany these structural changes [42]. This gap is critical because pathogenic invasion and biofilm resilience are governed by the intricate relationship between surface phenomena and intracellular responses.
Correlative microscopy addresses this fundamental challenge by integrating HS-AFM with optical techniques that report on internal cellular states. This powerful combination allows researchers to link nanoscale surface dynamics, such as the activity of appendages or changes in membrane stiffness, with simultaneous intracellular events occurring within the same cell [42] [43]. For researchers investigating dynamic biofilm processes, this correlation is essential to move beyond descriptive morphology to a mechanistic understanding of how structural changes relate to functional outcomes, such as antimicrobial resistance or host-pathogen interactions. The following sections provide a detailed framework for implementing this approach, from experimental design to data synthesis.
Integrated Workflow for Correlative HS-AFM and Fluorescence Microscopy
The following diagram outlines the core experimental workflow for conducting a correlative microscopy study of biofilm dynamics, from sample preparation through final data correlation.
Key Methodologies and Experimental Protocols
Probe Functionalization for Force Spectroscopy and Ligand Delivery
A critical advantage of AFM in biofilm research is its ability to function as a precise mechanical tool, not just an imaging device. Functionalizing the AFM cantilever with specific biomolecules enables the investigation of receptor-ligand interactions, adhesion forces, and even targeted stimulation of individual bacterial cells. The protocol below details the attachment of stimulatory molecules (e.g., antibodies, peptide-MHC) to the cantilever tip for subsequent surface receptor ligation on microbial or host cells [44].
Basic Protocol: Covalent Attachment of Biomolecules to AFM Cantilevers
Materials:
- AFM cantilevers (e.g., HYDRA6R-200N from AppNano)
- (3-Mercaptopropyl)-trimethoxysilane
- Streptavidin
- Sulfo-LC-SPDP (Sulfosuccinimidyl 6-(3'-(2-pyridyldithio)propionamido)hexanoate)
- Biotinylated molecule of interest (e.g., anti-CD3 antibody, adhesive protein)
- Poly-dimethylsiloxane (PDMS)
- Plasma cleaner
- Vacuum oven
- Zeba Desalting spin columns
Step-by-Step Procedure:
- Cantilever Preparation: Measure the spring constant of each cantilever using the thermal noise method. Plasma-clean the cantilevers for one minute to activate the surface.
- Silanization: Submerge the plasma-cleaned cantilevers in acetone containing 4% (3-Mercaptopropyl)-trimethoxysilane for 2 hours. This forms a reactive thiol-terminated layer.
- Washing and Curing: Wash the cantilevers sequentially with excess acetone, isopropanol, and distilled deionized water. Cure them in a vacuum oven at 110 °C for 1 hour.
- Streptavidin Functionalization:
- Prepare a 20 mM solution of sulfo-LC-SPDP in ultrapure water.
- React 3.125 µL of this solution with 250 µg of streptavidin in 125 µL of PBS containing 1 mM EDTA.
- Remove unreacted cross-linker using a Zeba Desalting spin column, equilibrated with PBS-EDTA.
- Coating: Pipette 25 µL of the Streptavidin-sulfo-LC-SPDP solution onto each silanized cantilever and allow the reaction to proceed overnight at 4 °C in a humidified chamber.
- Final Assembly: Wash the cantilevers three times with PBS to remove unbound streptavidin. The cantilever is now ready for the attachment of any biotinylated ligand by simple incubation [44].
Large-Area HS-AFM for Mapping Biofilm Heterogeneity
A major challenge in conventional AFM is the limited field of view, which can miss the inherent spatial heterogeneity of biofilms. The following protocol, adapted from recent work, describes an automated large-area AFM approach capable of capturing high-resolution images over millimeter-scale areas, which is essential for linking local cellular events to the architecture of the broader microbial community [2].
Application Note: Automated Large-Area HS-AFM of Biofilms
- Objective: To map the spatial heterogeneity and cellular morphology during the early stages of biofilm formation over large, biologically relevant areas.
- Sample Preparation: Grow biofilms (e.g., Pantoea sp. YR343) on treated glass or silicon substrates (e.g., PFOTS-treated coverslips) in a petri dish. At selected time points, remove the substrate, gently rinse to remove unattached cells, and air-dry before imaging [2].
- Instrumentation Setup:
- Employ an AFM system with a large-range piezoelectric scanner (capable of >100 µm motion).
- Implement an automated stage to move the sample between adjacent imaging regions.
- Use a closed-loop scanner to minimize positional drift and piezo creep over long acquisitions.
- Data Acquisition:
- Define a grid of multiple, adjacent scanning positions over the desired millimeter-scale area.
- Acquire high-resolution AFM topographs at each position with minimal overlap (5-10%).
- The process can be automated to run for extended periods (multiple days) with minimal user intervention [2].
- Data Processing and Analysis:
- Image Stitching: Use machine learning (ML)-aided algorithms to seamlessly stitch individual topographs into a single, large-area map, even with limited overlapping features.
- Automated Analysis: Apply ML-based image segmentation to the stitched image for automated cell detection, classification, and extraction of quantitative parameters (e.g., cell count, confluency, cellular orientation, filamentation) [2].
Table 1: Quantitative Data from Large-Area AFM of Pantoea sp. YR343 Biofilm [2]
| Parameter | Value | Significance |
|---|---|---|
| Individual Cell Length | ~2 µm | Corresponds to a surface area of ~2 µm²; aligns with known morphology. |
| Individual Cell Diameter | ~1 µm | - |
| Flagellar Height | ~20-50 nm | Confirms identity as flagella; visualized bridging gaps between cells. |
| Cluster Pattern | Honeycomb-like gaps | Emergent architecture observed after 6-8 hours of surface growth. |
| Field of View | Millimeter-scale | Enables observation of macroscale spatial organization previously obscured. |
The Scientist's Toolkit: Essential Reagents and Materials
Successful implementation of correlative microscopy requires careful selection of reagents and materials. The following table details key solutions for the protocols described in this note.
Table 2: Research Reagent Solutions for Correlative Microscopy
| Item | Function/Application | Key Details & Examples |
|---|---|---|
| Functionalized Cantilevers | Force spectroscopy, ligand delivery, molecular recognition. | Silanized tips with covalently bound streptavidin for biotinylated ligand attachment (e.g., antibodies, adhesive proteins) [44]. |
| Optically Compatible Substrates | Biofilm growth for simultaneous AFM and optical imaging. | PFOTS-treated glass coverslips; silicon wafers with modified surface chemistry to control bacterial adhesion [2]. |
| Live-Cell Fluorescent Dyes | Reporting internal cellular events in real-time. | Calcium indicators (e.g., Fluo-4), viability stains (e.g., SYTO 9/propidium iodide), GFP-transfected cells for protein localization [42]. |
| Cross-linking Reagents | Covalent attachment of molecules to AFM probes. | Heterobifunctional cross-linkers like Sulfo-LC-SPDP, which link surface amines on proteins to thiolated cantilevers [44]. |
| Microfluidic Cultivation Systems | Long-term, in-situ imaging under controlled conditions. | Enables continuous nutrient flow and waste removal, allowing visualization of biofilm development dynamics [45]. |
Data Correlation and Integration Framework
The final and most critical phase is the integrated analysis of multi-modal datasets. The diagram below illustrates the logical pathway for correlating data from different microscopy sources to build a comprehensive model of biofilm activity.
Analytical Protocol: Data Synthesis
Spatial Registration:
- Use fiduciary markers or distinctive cellular landmarks present in both AFM and optical images to align the datasets precisely.
- Software such as PhotoPad Image Editor or Adobe Photoshop can be used for manual alignment, while specialized correlative microscopy platforms offer automated registration [46].
Temporal Synchronization:
- Synchronize the clocks of the HS-AFM and optical microscopy systems prior to experiment initiation.
- Timestamp each data frame from both modalities to construct a unified timeline of events.
Quantitative Parameter Extraction:
- From HS-AFM: Extract parameters like surface roughness, cellular dimensions, elastic (Young's) modulus, and adhesion force maps.
- From Optical Microscopy: Quantify fluorescence intensity, its spatial distribution, and temporal kinetics (e.g., calcium spikes).
Model Building:
- Statistically analyze the correlation between nanomechanical events (e.g., local softening of the cell membrane) and the onset of an internal biochemical signal (e.g., a fluorescence intensity change).
- This allows researchers to move from observation to causation, building testable models of structure-function relationships in dynamic biofilm processes [42] [43].
The investigation of dynamic biofilm processes necessitates analytical techniques that can resolve both structural features and their underlying mechanical properties. While high-speed atomic force microscopy (AFM) imaging provides exceptional topographical details of biofilm formation and architecture, these structural observations require validation through quantitative mechanical measurements. Quantitative nanomechanical mapping (QNM) has emerged as a pivotal methodology that bridges this analytical gap by enabling simultaneous topographical imaging and spatial mapping of mechanical properties at the nanoscale. When integrated with force spectroscopy, researchers gain a powerful validation framework to correlate structural observations with quantitative mechanical data, providing unprecedented insights into biofilm development, resilience, and response to therapeutic interventions.
The significance of this integrated approach is particularly evident in biofilm research, where mechanical properties directly influence fundamental biological behaviors and treatment outcomes. Biofilm mechanical characteristics, including elasticity, adhesion, and viscoelasticity, govern bacterial adhesion strength, structural integrity, and resistance to mechanical disruption [4]. Furthermore, these properties significantly impact antimicrobial penetration and efficacy, making their accurate quantification essential for developing effective anti-biofilm strategies [3]. This application note details protocols for utilizing force spectroscopy to validate structural observations within the context of high-speed AFM imaging of dynamic biofilm processes, providing researchers with methodological frameworks to enhance the quantitative rigor of their investigations.
Technical Comparison of AFM Modalities
Atomic force microscopy offers multiple operational modes for investigating biological samples, each with distinct advantages and limitations for biofilm research. Understanding these distinctions is crucial for selecting the appropriate methodology for integrating structural and mechanical characterization.
Table 1: Comparison of AFM Nanomechanical Characterization Techniques
| Technique | Maximum Speed | Force Control | Key Advantages | Primary Limitations |
|---|---|---|---|---|
| Traditional Force Spectroscopy | ~100 Hz [47] | Moderate | Constant loading/unloading rates; well-established models [47] [48] | Slow acquisition; limited spatial mapping capability |
| Force Volume | ~10 Hz [48] | Moderate | Direct comparison between topography and property maps; well-suited for viscoelastic studies [48] | Time-consuming (minutes to hours); potential sample drift |
| PeakForce QNM | ~1,000 Hz [48] | Excellent (as low as 10 pN) [48] | High-speed quantitative mapping; superior force control; reduced tip wear [48] | Less control over ramp parameters; requires specialized instrumentation |
| Photothermal PORT | >25 kHz [47] | Good | Ultra-high-speed mapping; reduced sample disturbance; gentle imaging [47] | Complex thermomechanical calibration; newer methodology |
The selection of an appropriate AFM modality depends heavily on specific research objectives. For high-resolution mapping of heterogeneous biofilm surfaces at cellular and subcellular scales, PeakForce QNM provides an optimal balance of speed, resolution, and quantitative accuracy [48]. When investigating time-dependent mechanical behaviors or kinetic processes, force volume remains valuable despite its slower acquisition speed [48]. For the most demanding dynamic studies requiring the highest temporal resolution, emerging technologies like photothermal off-resonance tapping (PORT) offer acquisition rates at least an order of magnitude faster than conventional methods [47].
Protocol: High-Speed Nanomechanical Mapping of Biofilms
This protocol describes the procedure for correlated structural and mechanical characterization of live biofilms using high-speed AFM modalities, particularly PeakForce QNM and photothermal PORT.
Sample Preparation and Immobilization
Substrate Selection and Functionalization:
- Use glass coverslips treated with poly-L-lysine or PFOTS (perfluorooctyltrichlorosilane) to promote bacterial adhesion while maintaining viability [2] [4].
- For difficult-to-immobilize specimens, consider patterned polydimethylsiloxane (PDMS) stamps with microstructures tailored to cell dimensions (1.5-6 µm wide, 1-4 µm depth) [4].
Biofilm Growth and Stabilization:
- Inoculate surfaces with bacterial suspension (e.g., Pantoea sp. YR343) in appropriate growth medium [2].
- Incubate under optimal conditions for desired biofilm development stage (30 min for initial attachment studies; 6-8 hours for microcolony formation) [2].
- Gently rinse with appropriate buffer (e.g., PBS) to remove non-adherent cells, preserving the intact biofilm structure [2].
- For live-cell imaging, maintain hydrated conditions throughout transfer to AFM fluid cell.
AFM Instrument Configuration
Probe Selection:
- For soft biological samples (E < 100 kPa), use cantilevers with spring constants of 0.06-0.08 N/m [49].
- Select appropriate tip geometry:
System Calibration:
Measurement Parameters:
Data Acquisition and Processing
Topographical and Mechanical Mapping:
- Engage tip with surface using standard engagement procedure.
- Simultaneously acquire height, adhesion, deformation, and modulus channels.
- For large-area analysis, implement automated tile scanning with 10-15% overlap [2].
- Apply real-time feedback gains to maintain stable tip-sample interaction while minimizing imaging force.
Force Curve Processing:
Protocol: Force Spectroscopy for Structural Validation
This protocol provides a framework for using single-point and mapping force spectroscopy to validate structural features observed in high-speed AFM imaging of biofilms.
Targeted Force Mapping
Region of Interest Identification:
Spatially-Correlated Force Measurement:
- Program force curve acquisition at specific coordinates corresponding to structural features.
- For cellular-level analysis, implement 5×5 to 10×10 force curve arrays over individual cells.
- Set approach/retract velocity to 20 µm/s for soft materials [49].
- Include sufficient hold time (0.1-1 s) at maximum force to assess viscoelastic relaxation where relevant.
Data Analysis and Correlation
Mechanical Property Extraction:
- Process individual force curves to determine local Young's modulus, adhesion, and deformation.
- For heterogeneous features like bacterial honeycomb patterns [2], calculate average properties for structurally similar regions.
- Generate modulus distribution histograms to quantify property heterogeneity.
Structure-Property Correlation:
- Create overlay maps of topography and mechanical properties.
- Perform statistical analysis to establish correlation between structural height features and mechanical measurements.
- Calculate property gradients at structural boundaries (e.g., cell-cell interfaces, matrix-cell boundaries).
Table 2: Representative Nanomechanical Properties of Biofilm Components
| Biofilm Component | Young's Modulus Range | Adhesion Force | Measurement Considerations |
|---|---|---|---|
| Bacterial Cell Body | 0.5-15 MPa [48] [49] | 0.5-2 nN | Significant variation during division (2 vs. 15 MPa) [48] |
| Extracellular Matrix | 0.1-2 kPa [49] | 1-5 nN | Highly dependent on hydration and composition |
| Flagellar Structures | ~2 MPa [48] | Not reported | Require high spatial resolution to resolve 20-50 nm structures [2] |
| Model Hydrogels | 100 Pa-10 kPa [49] | Minimal | Used as calibration standards for soft measurements |
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful implementation of quantitative nanomechanical mapping for biofilm validation requires specific materials and reagents optimized for soft biological measurements.
Table 3: Essential Research Reagents and Materials for Biofilm QNM
| Item | Specifications | Function | Application Notes |
|---|---|---|---|
| AFM Probes | Sharp pyramidal tips (MLCT, Bruker); k ≈ 0.08 N/m [49] | High-resolution topography and mechanics | Cone semi-angle 17°; optimal for Sneddon model fitting |
| Colloidal Probes | Spherical tips; R = 2.5 µm; k ≈ 0.06 N/m [49] | Reduced local pressure; improved model applicability | Superior for very soft samples (E < 1 kPa); minimal sample damage |
| Calibration Samples | PDMS standards; PNIPAM hydrogels (100 Pa-10 kPa) [49] | System validation and method optimization | Essential for soft sample measurements; establish accuracy limits |
| Functionalized Substrates | PFOTS-treated glass; poly-L-lysine coatings [2] [4] | Sample immobilization | Promote adhesion while maintaining viability |
| Immobilization Aids | Patterned PDMS stamps; microstructures (1.5-6 µm) [4] | Secure positioning of recalcitrant cells | Enable oriented immobilization for consistent measurements |
| Liquid Cells | Temperature-controlled fluid chambers | Maintenance of physiological conditions | Preserve biofilm viability during extended measurements |
Data Interpretation and Validation Framework
The integration of structural and mechanical data requires a systematic approach to ensure accurate biological interpretation and methodological validation.
Analytical Workflow for Correlation Validation
Quality Assessment of Force Curves:
- Evaluate force curves for consistent contact point detection, which is particularly challenging in liquid environments [49].
- Exclude curves exhibiting excessive noise, irregular baselines, or inconsistent contact regions.
- Verify that indentation depth does not exceed 10-20% of sample thickness to avoid substrate effects [49].
Model Selection and Applicability:
- Select contact mechanics models based on tip geometry and sample properties:
- Validate model applicability through fitting quality assessment and comparison with reference materials.
Spatial Correlation Analysis:
- Register mechanical property maps with topographical features using cross-correlation algorithms.
- Quantify property differences between structurally distinct regions using statistical tests (e.g., t-tests for cell body vs. matrix).
- Calculate property gradients at interfaces to understand mechanical transitions.
Methodological Cross-Validation
Reference Material Validation:
Multi-Technique Corroboration:
The integration of quantitative nanomechanical mapping with force spectroscopy provides a robust analytical framework for validating structural observations in biofilm research. The protocols detailed in this application note enable researchers to move beyond purely descriptive topological characterization to obtain quantitative mechanical insights that illuminate structure-function relationships in microbial systems. As AFM technologies continue to advance, particularly with the development of high-speed modalities like photothermal PORT and automated large-area mapping, the capacity to correlate dynamic structural changes with spatiotemporal mechanical properties will dramatically enhance our understanding of biofilm development, resilience, and response to therapeutic intervention. This methodological approach establishes a foundation for truly quantitative analysis of biofilm systems, with significant implications for both fundamental microbiology and applied therapeutic development.
The study of biofilm mechanics is pivotal for advancing our understanding of microbial communities, from initial adhesion to mature biofilm formation and dispersal. However, the biofilm research community currently faces a significant challenge: the lack of standardized protocols for mechanical characterization. This absence leads to method-dependent results, with reported mechanical property values often varying by several orders of magnitude for the same bacterial strain [50]. Such variability hinders reliable comparison of microbiological protocols essential for improving engineering processes and antimicrobial screening [50]. Within the broader context of research on high-speed Atomic Force Microscopy (AFM) imaging of dynamic biofilm processes, this application note addresses the critical need for standardized methodologies. We present detailed protocols and analytical frameworks to enhance reproducibility in biofilm mechanical characterization, enabling researchers and drug development professionals to generate comparable, high-quality data across laboratories and experimental setups.
The Imperative for Standardization in Biofilm Mechanics
Biofilms, defined as complex three-dimensional microbial communities embedded in a self-produced extracellular polymeric substance (EPS), exhibit mechanical properties that are crucial to their function and persistence [50] [33]. The mechanical behavior of a biofilm—how it responds to external forces—directly influences its physical stability, dissemination potential, and resistance to mechanical and chemical stresses [50]. Understanding these properties is therefore essential for developing effective anti-biofilm strategies in medical contexts and for optimizing beneficial biofilm applications in biotechnological processes.
The current landscape of biofilm mechanical characterization is marked by a pronounced lack of consensus. Research shows that "literature values often differ by several orders of magnitude for the same bacterial strain" due to methodological differences [50]. This variability stems from several factors:
- Diverse mechanical testing methods (e.g., AFM, rheometry, microindentation)
- Varying identification methods for mechanical parameters
- Sample-to-sample heterogeneity inherent to living biofilm structures
- Differences in cultivation conditions and experimental setups
This methodological diversity creates a critical "blind spot in the biofilm community" that compromises the reliability of comparative studies, particularly for antibiotic screening and the development of anti-biofilm therapeutics [50]. Initiatives like MIABiE (Minimum Information About a Biofilm Experiment) have emerged to address these challenges by providing guidelines for the minimum information required to document and store biofilm experiments [50] [51]. Similarly, the International Biofilm Standards Tasks Group (IBSTG) works to coordinate global standardization efforts [52]. These frameworks aim not to prescribe how experiments should be performed, but rather to establish what information must be reported to ensure interpretability and independent verification of results [51].
Mechanical Properties of Biofilms: Significance and Parameters
The Microbiological Significance of Biofilm Mechanics
Mechanical parameters provide crucial insights into biofilm behavior with far-reaching implications:
- Understanding Biofilm Life-Cycle: Mechanical properties influence the transition from initial attachment to microcolony formation and eventual dispersal. Environmental stimuli such as fluid flow and shear stress actively shape biofilm architecture by affecting exopolysaccharide production and cyclic di-GMP signaling [50].
- Viscoelastic Behavior: Biofilms are viscoelastic materials, capable of both dissipating energy (viscous response) and recovering their shape (elastic response) when subjected to mechanical stress. This property determines how biofilms withstand external perturbations and form problematic structures like streamers that clog medical devices [50].
- Antimicrobial Screening: Changes in mechanical properties serve as biomarkers for evaluating antibiofilm treatment efficacy. Treatments that target EPS components can reduce biofilm cohesiveness or stiffness, potentially enhancing biocide penetration and effectiveness [50]. Mechanical parameters thus provide quantitative measures of treatment impact beyond simple viability counts.
Key Mechanical Parameters and Their Interpretation
The following table summarizes the fundamental mechanical properties relevant to biofilm characterization:
Table 1: Key Mechanical Properties in Biofilm Characterization
| Mechanical Property | Definition | Biological Significance | Common Measurement Techniques |
|---|---|---|---|
| Elasticity (Young's Modulus) | Resistance to reversible deformation under stress | Indicates structural rigidity and matrix integrity; stiffer biofilms may resist detachment | AFM force spectroscopy, microindentation [50] [53] |
| Adhesion | Force required to separate biofilm from substrate or internal cohesion | Determines colonization potential and cleanability of surfaces | AFM retraction curve analysis, centrifugation assays [53] |
| Viscoelasticity | Combination of viscous (liquid-like) and elastic (solid-like) responses | Influences deformation and recovery under fluid flow; critical for dispersal | Rheometry, creep testing [50] |
| Cohesiveness | Internal strength binding biofilm components | Affects stability and fragment detachment potential | Tensile testing, micromanipulation [50] |
Standardized Protocols for AFM-Based Mechanical Characterization
Atomic Force Microscopy offers unique capabilities for quantifying mechanical properties at the nanoscale under physiologically relevant conditions [53]. The following protocols establish standardized approaches for AFM-based mechanical characterization of biofilms.
Biofilm Cultivation and Sample Preparation
Principle: Reproducible mechanical characterization begins with standardized biofilm growth conditions. The static microtiter plate assay provides a high-throughput method for consistent biofilm formation [54].
Materials:
- 96-well microtiter plates, not tissue culture-treated (e.g., Becton Dickinson #353911)
- Appropriate growth medium for target microorganisms
- Sterile 96-prong inoculating manifold (for high-throughput studies)
- Crystal violet solution (0.1% w/v in water) for quality control
- Acetic acid (30% v/v) or other appropriate solvent for dye elution
Procedure:
- Inoculate bacterial strains in 3-5 mL culture and grow to stationary phase.
- Dilute cultures 1:100 in fresh medium.
- Pipette 100 μL of diluted culture into quadruplicate wells of a microtiter plate.
- Cover plate and incubate at optimal growth temperature for defined time (typically 24-48 hours, determined empirically for each strain).
- For quality control, include crystal violet staining to confirm consistent biofilm formation across replicates [54].
Critical Considerations:
- Maintain consistent lid usage: clean lids with 70% ethanol between experiments
- Document temperature, medium composition, and incubation time precisely
- For high-throughput screens, use direct inoculation from overnight microtiter plate cultures with a sterile 96-prong manifold [54]
Sample Immobilization for AFM Analysis
Principle: Proper immobilization is essential for reliable AFM measurements. The method must secure biofilms without altering their native mechanical properties [53].
Materials:
- Glass coverslips or specialized AFM substrates
- Poly-L-lysine solution or Corning Cell-Tak
- Polydimethylsiloxane (PDMS) stamps or polycarbonate membranes (for yeast cells)
- Phosphate-buffered saline (PBS) for washing
Procedure:
- Surface Treatment: Apply poly-L-lysine or Cell-Tak to create a positively charged surface according to manufacturer specifications.
- Biofilm Transfer: Carefully extract biofilms from growth substrate and place on treated surface.
- Secure Immobilization: For robust attachment, use PDMS stamps or polycarbonate membranes to physically trap cells without chemical modification.
- Washing: Gently rinse with PBS to remove non-adherent cells while preserving biofilm integrity.
Critical Considerations:
- Chemical fixatives should be avoided as they alter mechanical properties
- The EPS matrix of native biofilms may provide sufficient adhesion without additional treatments
- Document immobilization method thoroughly, including product lots and concentrations [53]
AFM Force Spectroscopy and Data Acquisition
Principle: AFM force spectroscopy measures mechanical properties by monitoring tip-sample interactions during approach-retraction cycles [53].
Materials:
- Atomic Force Microscope with fluid cell capability
- Appropriate cantilevers (typical spring constants: 0.01-0.1 N/m for biofilms)
- Calibration substrates (e.g., clean glass slide)
- Liquid medium compatible with biofilm viability
Procedure:
- Cantilever Calibration: Determine the spring constant (k_cantilever) using thermal tuning or reference measurements on a hard surface in fluid [53].
- Experimental Setup: Engage fluid cell and position cantilever above biofilm sample.
- Force Curve Acquisition:
- Program approach-retraction cycles with specified ramp size (typically 1-5 μm)
- Set approach/retraction speeds (typically 0.5-2 μm/s)
- Define trigger threshold to prevent sample damage
- Acquire multiple curves (≥100) across different sample regions
- Parameter Settings:
- Approach distance: Sufficient to establish contact and compress sample
- Dwell time: 0-1 second at maximum compression
- Retraction distance: Sufficient to break all adhesive interactions
Critical Considerations:
- Perform measurements in liquid to eliminate capillary forces
- Maintain consistent loading rates across experiments
- Validate set point force to avoid sample damage
- Document all instrument parameters including cantilever type, spring constant, and approach/retraction speeds [53]
Data Analysis and Interpretation
Principle: Mechanical parameters are extracted from force-distance curves through appropriate physical models.
Analysis Workflow:
- Approach Curve Analysis:
- Elastic Modulus: Fit nonlinear compression region with Hertz model or its variations
- Cell Stiffness: Calculate from slope of linear compression region using spring series model: 1/keffective = 1/kcell + 1/k_cantilever [53]
- Retraction Curve Analysis:
- Adhesion Force: Measure maximum force required to separate tip from sample
- Adhesion Work: Calculate area under retraction curve
- Tether Formation: Note discontinuities indicating membrane tethers or polymer unfolding
Critical Considerations:
- Apply consistent fitting parameters across data sets
- Report model assumptions and limitations
- Include sample size (number of curves) and statistical measures
- Acknowledge potential artifacts from surface heterogeneity [53]
Advanced Approaches: Large Area AFM and Machine Learning Integration
Traditional AFM faces limitations in studying biofilms due to its restricted scan range (<100 μm), which struggles to capture the spatial heterogeneity of millimeter-scale biofilm structures [2]. Recent advances address this challenge through automated large area AFM coupled with machine learning.
Large Area AFM Methodology
Principle: Automated large area AFM combines multiple high-resolution scans to create comprehensive maps of biofilm topography and mechanical properties across millimeter scales [2].
Implementation:
- Hardware Requirements: AFM system with large-range piezoelectric actuators and automated stage
- Image Acquisition: Program sequential tile scanning with minimal overlap (5-10%)
- Image Stitching: Apply algorithms to seamlessly combine individual scans
- Data Integration: Correlate AFM data with complementary techniques (e.g., fluorescence microscopy)
Applications:
- Visualization of cellular orientation patterns (e.g., honeycomb structures in Pantoea sp. YR343) [2]
- Mapping of flagellar interactions and EPS distribution during early biofilm development
- Correlation of local mechanical properties with structural features
Machine Learning-Enhanced Analysis
Principle: Machine learning transforms AFM data acquisition and analysis through four key areas: sample region selection, scanning process optimization, data analysis, and virtual AFM simulation [2].
Implementation:
- Image Analysis:
- Automated cell detection and classification
- Morphological parameter extraction (cell count, confluency, shape, orientation)
- Identification of nanoscale features (flagella, pili, EPS fibers)
- Scanning Optimization:
- AI-driven region selection to target representative areas
- Sparse scanning approaches with ML-based image reconstruction
- Automated probe conditioning and health monitoring
Benefits:
- Enhanced statistical power through analysis of thousands of cells
- Elimination of operator bias in feature identification
- Increased throughput through automated operation [2]
The following diagram illustrates the integrated workflow for advanced AFM analysis of biofilms:
Diagram 1: Integrated workflow for advanced AFM analysis of biofilms combining large-area scanning with machine learning.
Essential Research Reagent Solutions
Successful mechanical characterization of biofilms requires specific materials and reagents. The following table details essential components and their functions:
Table 2: Essential Research Reagents for Biofilm Mechanical Characterization
| Reagent/Material | Function | Application Notes | References |
|---|---|---|---|
| Poly-L-lysine | Surface coating for cell immobilization | Creates positive charge for electrostatic attachment; suitable for many bacterial species | [53] |
| Corning Cell-Tak | Enhanced surface adhesion | Provides more robust attachment for force spectroscopy; recommended for difficult-to-immobilize cells | [53] |
| PDMS Stamps | Physical entrapment of cells | Alternative to chemical fixation; maintains native mechanical properties | [53] |
| Crystal Violet (0.1%) | Biofilm biomass quantification | Quality control for consistent biofilm formation across samples | [54] |
| Acetic Acid (30%) | Dye solubilization | Efficiently elutes crystal violet for spectrophotometric quantification | [54] |
| PFOTS-treated Glass | Hydrophobic surface modification | Controls bacterial adhesion patterns for standardized initial attachment studies | [2] |
Data Reporting Standards and Minimum Information Guidelines
Consistent reporting following established guidelines is essential for experimental reproducibility and data comparison across studies.
MIABiE Reporting Framework
The Minimum Information About a Biofilm Experiment (MIABiE) initiative provides a modular framework for comprehensive documentation of biofilm studies [51]. Key reporting modules relevant to mechanical characterization include:
- Sample Generation and Study Design: Document microbial strains, environmental conditions, and analytical technologies
- Biofilm Model System: Specify reactor type (e.g., flow cell, microtiter plate) and operational parameters
- Analytical Techniques: Detail mechanical testing methods and associated parameters
- Data Analysis: Describe processing methods, statistical approaches, and quality controls
Essential Metadata for Mechanical Characterization
The following diagram outlines the critical parameters that must be documented for reproducible mechanical characterization:
Diagram 2: Critical parameters for documentation in biofilm mechanical characterization studies.
Standardized protocols for mechanical characterization of biofilms are essential for advancing both fundamental understanding and applied interventions. The integration of traditional AFM methodologies with emerging technologies such as large-area scanning and machine learning represents a promising path toward comprehensive, multi-scale analysis of biofilm mechanical properties. By adopting the standardized protocols, reporting frameworks, and analytical approaches outlined in this application note, researchers can enhance the reproducibility, reliability, and comparability of biofilm mechanical data. Such standardization will accelerate progress in antimicrobial development, biofilm-based bioprocess optimization, and the fundamental understanding of microbial community mechanics. As the field evolves, continued collaboration between academic researchers, industry professionals, and standards organizations will be crucial for refining these protocols and addressing new challenges in biofilm mechanobiology.
Conclusion
High-Speed AFM has unequivocally emerged as a pivotal technology for demystifying the dynamic world of biofilms, providing an unprecedented real-time view of their assembly, architecture, and functional adaptations. By bridging the critical gap between nanoscale cellular events and the emergent macroscale properties of the biofilm community, HS-AFM offers profound insights. The integration of automation and machine learning is set to further revolutionize this field, enabling high-throughput screening and deeper quantitative analysis. For biomedical research and drug development, these advances pave the way for rationally designed interventions that target biofilm cohesion, resilience, and dispersal mechanisms, ultimately leading to more effective therapies against persistent biofilm-associated infections and innovative applications in biotechnology.
References
- 1. Artificial intelligence-empowered imaging technologies for ... [https://www.sciencedirect.com/science/article/abs/pii/S1385894725084311]
- 2. Analysis of biofilm assembly by large area automated AFM [https://www.nature.com/articles/s41522-025-00704-y]
- 3. Microscopy Methods for Biofilm Imaging: Focus on SEM ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7828176/]
- 4. Atomic Force Microscopy of Biofilms—Imaging, Interactions ... [https://www.intechopen.com/chapters/50739]
- 5. A high signal-to-noise ratio and high-frequency seesaw ... [https://www.nature.com/articles/s41467-025-65240-x]
- 6. Nonlinear Modeling of a Piezoelectric Actuator-Driven High ... [https://www.mdpi.com/2076-0825/13/10/391]
- 7. Atomic Force Microscopes for Materials Research [https://www.bruker.com/en/products-and-solutions/microscopes/materials-afm.html]
- 8. Highâ€Speed Atomic Force Microscopy Reveals the Dynamic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12412613/]
- 9. Analysis of biofilm assembly by large area automated AFM [https://pmc.ncbi.nlm.nih.gov/articles/PMC12062311/]
- 10. Probing the reduction of adhesion forces between biofilms ... [https://www.sciencedirect.com/science/article/abs/pii/S0927776523005957]
- 11. Technical advances in high-speed atomic force microscopy [https://pmc.ncbi.nlm.nih.gov/articles/PMC10771405/]
- 12. - High Advancements for Protein Speed Research AFM Dynamics [https://www.azonano.com/article.aspx?ArticleID=6725]
- 13. Real-Time Monitoring of Biofilm Formation Using a ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9853957/]
- 14. Applications of high - speed to real-time... atomic force microscopy [https://pubmed.ncbi.nlm.nih.gov/28716648/]
- 15. A guide for nanomechanical characterization of soft matter ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12166430/]
- 16. Advances in biofilm characterization: utilizing rheology and ... [https://link.springer.com/article/10.1007/s42114-024-00950-2]
- 17. PinPointâ„¢ Nanomechanical Mode [https://www.parksystems.com/en/products/research-afm/AFM-modes/Nanomechanical-Modes/pinpoint--afm-nanoelectrical-modes]
- 18. Advances in nanomechanical property mapping by atomic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12379807/]
- 19. Improving image contrast and material discrimination with ... [https://www.nature.com/articles/ncomms7270]
- 20. Biofilms Exposed: Innovative Imaging and Therapeutic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12466655/]
- 21. Escherichia coli for Immobilizing studies of surface dynamics live AFM [https://pubmed.ncbi.nlm.nih.gov/24286980/]
- 22. Bacterial Immobilization for Imaging by Atomic Force Microscopy [https://www.jove.com/t/2880/bacterial-immobilization-for-imaging-by-atomic-force-microscopy]
- 23. In-Situ Determination of the Mechanical Properties of... | PLOS One [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0061663]
- 24. Analysis of Biofilm Assembly by Large Area Automated AFM [https://www.nanosurf.com/en/en/analysis-of-biofilm-assembly-by-large-area-automated-afm]
- 25. High-speed atomic force microscopy [https://pmc.ncbi.nlm.nih.gov/articles/PMC10771478/]
- 26. Machine learning assisted classification of staphylococcal ... [https://www.sciencedirect.com/science/article/pii/S2590207525000310]
- 27. Automated Blood Cell Detection and Classification in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11719705/]
- 28. Microbial Cells Analysis by Atomic Force Microscopy [https://www.sciencedirect.com/science/article/abs/pii/B9780123918567000251]
- 29. Recent advances in the application of atomic force ... [https://www.sciencedirect.com/science/article/abs/pii/S1047847723000266]
- 30. High-speed atomic force microscopy [https://pubmed.ncbi.nlm.nih.gov/23291302/]
- 31. AFM BioMed Conference 2025 - IBEC Events [https://events.ibecbarcelona.eu/afm-biomed-2025/]
- 32. Absolute Quantitation of Bacterial Biofilm Adhesion and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2711294/]
- 33. Quantitative and Qualitative Assessment Methods for Biofilm ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6133255/]
- 34. Atomic Force Microsocopy: Key Unconventional Approach for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11603319/]
- 35. Correction of AFM data artifacts using a convolutional ... [https://www.sciencedirect.com/science/article/abs/pii/S0304399122001851]
- 36. Numerical Study of Hydrodynamic Forces for AFM ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5662066/]
- 37. Methods to probe the formation of biofilms: applications in ... [https://pubs.rsc.org/en/content/articlehtml/2020/ay/c9ay02214g]
- 38. Understanding the Fundamental Basis for Biofilm ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2021.687118/full]
- 39. Molecular imaging of bacterial biofilms—a systematic review [https://pmc.ncbi.nlm.nih.gov/articles/PMC11523921/]
- 40. Microscopy Methods for Biofilm Imaging: Focus on SEM ... [https://www.mdpi.com/2079-7737/10/1/51]
- 41. Surface-enhanced Raman spectroscopy: a half-century ... [https://pubs.rsc.org/en/content/articlehtml/2025/cs/d4cs00883a]
- 42. AFM-Based Correlative Microscopy Illuminates Human ... [https://www.frontiersin.org/journals/cellular-and-infection-microbiology/articles/10.3389/fcimb.2021.655501/full]
- 43. Nanoscale monitoring of drug actions on cell membrane ... [https://www.nature.com/articles/aps201528]
- 44. Biochemical stimulation of immune cells and measurement of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6565469/]
- 45. Dynamics of bacterial biofilm development imaged using ... [https://pubmed.ncbi.nlm.nih.gov/40893774/]
- 46. A Step-By-Step Protocol for Correlative Light and Electron ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12336855/]
- 47. Highâ€Speed Quantitative Nanomechanical Mapping by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12508714/]
- 48. Toward Quantitative Nanomechanical Measurements on ... [https://www.bruker.com/en/products-and-solutions/microscopes/materials-afm/resource-library/an-141-toward-quantitative-nanomechanical-measurements-on-live-cells-with-peakforce-qnm.html]
- 49. Space-resolved quantitative mechanical measurements of ... [https://www.nature.com/articles/am2016170]
- 50. analysis and critical review | npj Biofilms and Microbiomes [https://www.nature.com/articles/s41522-018-0062-5]
- 51. Minimum information about a biofilm experiment (MIABiE) [https://pmc.ncbi.nlm.nih.gov/articles/PMC4406403/]
- 52. The role of standards in biofilm research and industry ... [https://www.sciencedirect.com/science/article/pii/S0964830522001603]
- 53. Using Atomic Force Microscopy To Illuminate the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7447269/]
- 54. Growing and Analyzing Static Biofilms - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4568995/]