AFM Nanomechanics in Biofilm Research: Decoding the Role of Capsular Polysaccharides
This article explores the pivotal role of Atomic Force Microscopy (AFM) nanomechanics in elucidating the structure-function relationship of capsular polysaccharides within bacterial biofilms.
AFM Nanomechanics in Biofilm Research: Decoding the Role of Capsular Polysaccharides
Abstract
This article explores the pivotal role of Atomic Force Microscopy (AFM) nanomechanics in elucidating the structure-function relationship of capsular polysaccharides within bacterial biofilms. Aimed at researchers, scientists, and drug development professionals, it synthesizes foundational knowledge with cutting-edge methodologies. The content covers the biomechanical principles of capsule-mediated adhesion, advanced AFM techniques for in situ analysis, strategies to overcome analytical challenges, and validation through comparative studies with other antibiofilm polysaccharides. By integrating the latest research, this review provides a comprehensive framework for leveraging AFM insights to develop novel anti-biofilm strategies, directly addressing the pressing challenge of antimicrobial resistance.
Capsular Polysaccharides and Biofilm Architecture: A Biomechanical Foundation
Bacterial biofilms represent a structured microbial community embedded within a self-produced extracellular polymeric substance (EPS) matrix. This complex matrix determines the physicochemical properties of biofilms and provides critical protection against environmental stresses, including antibiotics. Recent advances in atomic force microscopy (AFM) nanomechanics have enabled unprecedented high-resolution analysis of EPS components, particularly capsular polysaccharides, revealing new insights into their structural organization and functional properties. This technical guide examines biofilm architecture through the lens of AFM methodologies, providing researchers with foundational knowledge and experimental protocols for investigating the nanomechanical properties of EPS constituents in biofilm research.
The EPS Matrix: Composition and Functional Architecture
The extracellular polymeric substance (EPS) matrix establishes the functional and structural integrity of biofilms, constituting 50% to 90% of the total organic matter [1]. This matrix provides compositional support and protection for microbial communities in harsh environments [1]. Contrary to historical understanding, the EPS is a complex, dynamic assemblage of multiple biopolymer classes beyond polysaccharides.
Core Components of the EPS Matrix
The biofilm matrix is composed of several key macromolecular components, each contributing distinct functional properties:
Polysaccharides: Often heteropolymers containing neutral and charged sugar residues with organic/inorganic substituents [2]. Common examples include alginate in Pseudomonas aeruginosa biofilms, polysaccharide intercellular adhesion (PIA) in staphylococci, and cellulose in various environmental biofilms [3] [4].
Proteins: Including structural proteins that stabilize biofilm architecture and extracellular enzymes that facilitate nutrient acquisition and matrix remodeling [2]. Enzymes such as dispersin B, proteases, and DNases enable biofilm reorganization and dispersal [2].
Extracellular DNA (e-DNA): Provides structural integrity and facilitates genetic exchange [3]. In P. aeruginosa biofilms, e-DNA forms distinct grid-like structures and functions as an intercellular connector [3].
Lipids and Biosurfactants: Influence surface properties including wettability and charge, affecting bacterial adhesion and motility [2].
Membrane Vesicles: Act as "parcels" containing enzymes and nucleic acids, transported through the EPS matrix to participate in nutrient acquisition, gene exchange, and biological warfare [3].
Functional Classification of EPS Components
Table 1: Functional Classification of Major EPS Components Based on Neu and Lawrence's System [3]
| Function | EPS Component | Role in Biofilm |
|---|---|---|
| Constructive | Neutral polysaccharides, Amyloids | Structural framework and stability |
| Sorptive | Charged/hydrophobic polysaccharides | Ion exchange, sorption of dissolved substances |
| Active | Extracellular enzymes | Polymer degradation, nutrient acquisition |
| Surface-active | Amphiphilic compounds, Membrane vesicles | Interface interactions, export from cells |
| Informative | Lectins, Nucleic acids | Specificity recognition, genetic information |
| Redox active | Bacterial refractory polymers | Electron donor/acceptor functions |
| Nutritive | Various polymers | Source of carbon, nitrogen, phosphorus |
AFM Methodologies for EPS Nanomechanical Characterization
Atomic force microscopy has emerged as a powerful tool for investigating the structural and mechanical properties of biofilms at nanoscale resolution. Recent technological advances have addressed traditional limitations in imaging area and automation, enabling more comprehensive analysis of biofilm architecture.
Advanced AFM Imaging Techniques
Large Area Automated AFM: Traditional AFM imaging is limited to areas <100 μm, restricting analysis of heterogeneous biofilm structures. Recent developments combine automated large-area AFM with machine learning to capture high-resolution images over millimeter-scale areas [5]. This approach enables visualization of spatial heterogeneity and cellular morphology during early biofilm formation previously obscured by technical limitations [5].
Multiparametric PeakForce Tapping (PFT-AFM): This advanced mode enables high-resolution imaging of live bacteria under physiological conditions while simultaneously mapping nanomechanical properties [6]. The technique provides tight control over applied force (typically 1-6 nN), minimizing sample damage while allowing quantitative measurement of elastic modulus through Derjaguin-Muller-Toporov (DMT) models [6].
Experimental Protocol: AFM Analysis of Biofilm Mechanical Properties
Sample Preparation:
- Grow biofilms on appropriate substrates (e.g., PFOTS-treated glass coverslips) for selected time periods [5].
- Gently rinse to remove unattached cells while preserving EPS structure.
- For live cell imaging, immobilize bacteria in porous polycarbonate membranes to maintain physiological conditions [6].
AFM Imaging Parameters:
- Scanning Force: 1-6 nN peak force, optimized for EPS penetration while minimizing cellular damage [6].
- Scan Rate: 0.5-1.0 Hz, depending on required resolution and area.
- Resolution: 512 × 512 pixels for cellular-level analysis; higher resolutions (1024 × 1024) for subcellular features.
- Environment: Liquid phase for live cells; ambient conditions for fixed samples.
Data Analysis:
- Image Stitching: Apply machine learning algorithms to combine multiple high-resolution scans into seamless millimeter-scale images [5].
- Mechanical Mapping: Calculate elastic modulus (E) from force-distance curves using appropriate contact-mechanical models (DMT recommended) [6].
- Morphometric Analysis: Quantify cell dimensions, orientation, and spatial distribution using automated segmentation algorithms [5].
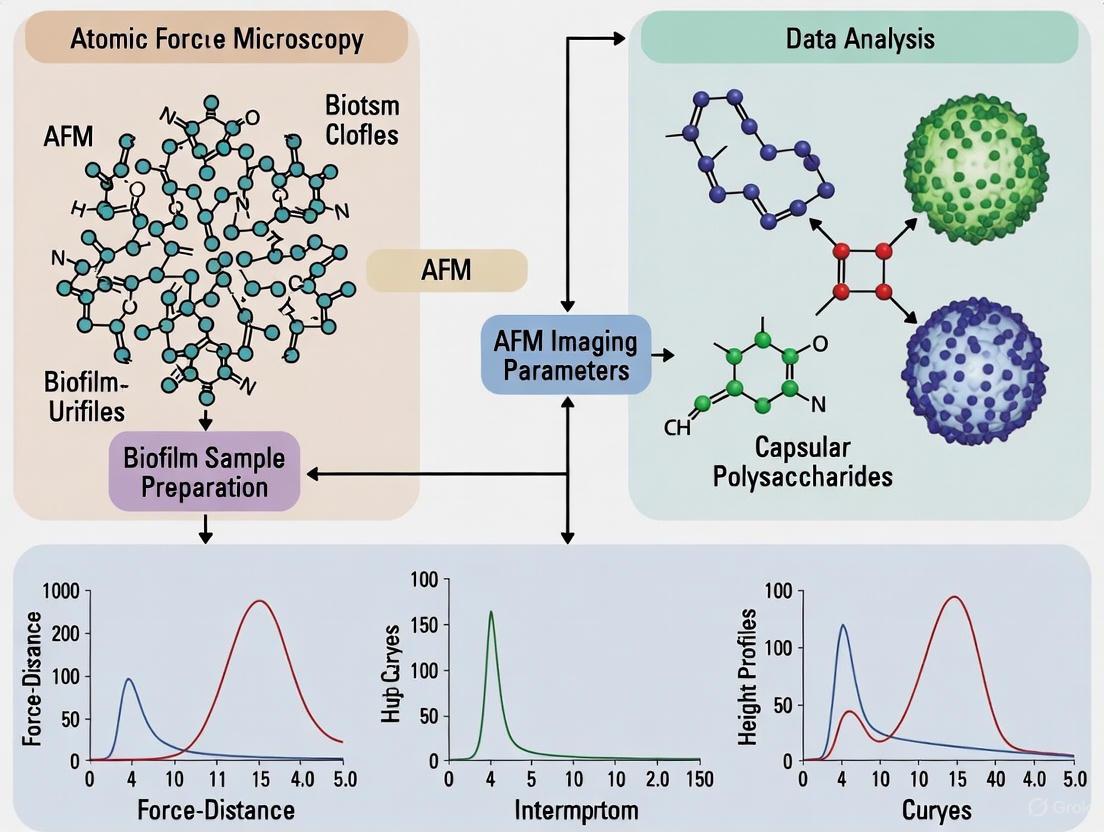
Figure 1: AFM Nanomechanics Workflow for Biofilm EPS Characterization. This diagram illustrates the integrated experimental and computational pipeline for analyzing the mechanical properties of EPS matrix components using advanced AFM methodologies.
Nanomechanical Properties of Capsular Polysaccharides
Capsular polysaccharides represent a critical EPS component with significant implications for biofilm mechanical properties and antibiotic resistance. AFM nanomechanics has revealed fundamental structure-function relationships in these biopolymers.
Structural Diversity of Bacterial Polysaccharides
Table 2: Common Bacterial Exopolysaccharides and Their Properties [4] [1]
| Polysaccharide | Producing Bacteria | Chemical Characteristics | Biofilm Function |
|---|---|---|---|
| Alginate | Pseudomonas spp., Azotobacter vinelandii | Polyanionic | Cell association, protection |
| Polysaccharide Intercellular Adhesion (PIA) | Staphylococcus aureus, S. epidermidis | Polycationic, β-1-6-linked N-acetylglucosamine | Cellular aggregation, adhesion |
| Cellulose | Acetobacter xylinum, Various bacteria | Neutral | Structural integrity, attachment |
| Pel | Pseudomonas aeruginosa | Neutral, composition unknown | Aggregation, pellicle formation |
| Xanthan | Xanthomonas campestris | Anionic heteropolymer | Structural stability |
| Hyaluronic acid | Streptococcus equi | Linear glycosaminoglycan | Adhesion, immune evasion |
Structure-Function Relationships in Antibiofilm Polysaccharides
Recent research has identified specific capsular polysaccharides with non-biocidal antibiofilm activity. Screening of 31 purified capsular polysaccharides revealed that active compounds share distinctive biophysical properties [7]:
- High Intrinsic Viscosity: Active polysaccharides demonstrate intrinsic viscosity >7 dl/g, significantly higher than inactive compounds [7].
- Molecular Weight Dependence: Size integrity is critical for antibiofilm activity; minor fragmentation of G2cps polysaccharide (average MW 800 kDa) resulted in complete loss of function [7].
- Electrokinetic Signature: Active macromolecules display distinct electrophoretic mobility under applied electric fields, characterized by high density of electrostatic charges and permeability to fluid flow [7].
Experimental Protocol: Screening Antibiofilm Polysaccharides
Polysaccharide Characterization:
- Purification: Isolate capsular polysaccharides from bacterial cultures using standard extraction protocols.
- Structural Analysis: Employ High-Performance Anion-Exchange Chromatography with Pulsed Amperometric Detection (HPAEC-PAD) and nuclear magnetic resonance (NMR) for composition analysis [7].
- Molecular Weight Determination: Utilize High Performance Size-Exclusion Chromatography coupled to Static Light Scattering (HPSEC-LS) to determine average molecular weight [7].
Biofilm Inhibition Assay:
- Static Microtiter Plate Assay: Incubate test bacteria with polysaccharides (typical concentration: 100 μg/mL) in 96-well plates [7].
- Crystal Violet Staining: Quantify biofilm biomass after appropriate incubation period.
- Dynamic Validation: Confirm results using continuous flow biofilm microfermentors under shear stress conditions [7].
- Viability Assessment: Perform colony counting to verify non-biocidal activity.
Research Reagent Solutions for EPS Characterization
Table 3: Essential Research Reagents for EPS and Biofilm Analysis
| Reagent/Category | Specific Examples | Research Function |
|---|---|---|
| Surface Substrates | PFOTS-treated glass, Polycarbonate membranes | Controlled surface attachment and AFM immobilization |
| Polysaccharide Standards | Vi polysaccharide, G2cps, PnPS3 | Positive controls for antibiofilm activity assays |
| Analytical Standards | Monosaccharide references, Molecular weight markers | Chromatographic calibration and structural validation |
| Enzymatic Tools | Dispersin B, Proteases, DNases | Selective EPS component degradation for functional studies |
| Staining Agents | Crystal violet, Fluorescent lectins | Biofilm visualization and quantification |
| Chromatography Media | HPAEC-PAD columns, HPSEC matrices | Polysaccharide separation and characterization |
EPS Matrix Architecture Visualized Through AFM
High-resolution AFM has revealed unprecedented details of biofilm EPS architecture, providing insights into the structural organization of matrix components:
Nanoscale Network Organization
Imaging of Pantoea sp. YR343 biofilms reveals a preferred cellular orientation with distinctive honeycomb patterning during early biofilm development [5]. AFM visualizes flagellar structures (20-50 nm in height) extending tens of micrometers across surfaces, forming bridging connections between cells [5].
In live Group B Streptococcus, multiparametric AFM reveals a net-like peptidoglycan architecture that stretches and stiffens in response to turgor pressure [6]. This network comprises parallel-oriented glycan strands that undergo structural rearrangement under osmotic stress, demonstrating the dynamic nature of matrix organization [6].
Matrix Architecture and Mechanical Response
Figure 2: EPS Matrix Response to Environmental Stimuli. This diagram illustrates the relationship between environmental cues, structural reorganization of EPS components, and resulting changes in mechanical properties that determine biofilm function.
Therapeutic Implications and Future Directions
The nanomechanical understanding of EPS components has significant implications for developing novel anti-biofilm strategies. Rather than traditional biocidal approaches, emerging interventions target the structural integrity of the EPS matrix:
- Matrix Disruption Therapies: Enzymatic degradation of key EPS components (e.g., DNases, dispersin B) can enhance antibiotic penetration [2].
- Non-biocidal Antiadhesion Agents: High molecular weight capsular polysaccharides with specific electrokinetic properties can prevent initial bacterial attachment without inducing resistance [7].
- Surface Engineering: Materials modified with antibiofilm polysaccharides can resist colonization in medical device applications [7] [2].
The integration of AFM nanomechanics with biochemical analysis provides a powerful framework for understanding structure-function relationships in biofilm EPS matrices, offering new avenues for controlling biofilm-associated infections in clinical settings.
Biofilms are structured communities of microbial cells embedded in a self-produced extracellular matrix, which confers significant tolerance to antimicrobial agents and host immune responses [8]. The polysaccharide components of this matrix are critical determinants of biofilm architecture, mechanics, and virulence. This whitepaper examines four key polysaccharides—PNAG, alginate, Psl, and Pel—within the context of atomic force microscopy (AFM) nanomechanics research. We provide a comprehensive technical analysis of their structural properties, functional roles, and biomechanical characteristics, supported by experimental data and methodologies relevant to drug development professionals seeking to disrupt pathogenic biofilms.
The biofilm matrix represents a critical interface between bacterial cells and their environment, strengthening the community structure while retaining mechanical plasticity [8]. Extracellular polymeric substances (EPS), particularly polysaccharides, typically constitute up to 85% of the biofilm volume and create heterogeneous local microenvironments that promote bacterial persistence [8]. The World Health Organization has classified antibiotic-resistant infections among its top 10 research priorities, with biofilm-related complications contributing significantly to the estimated 7 million annual deaths from antimicrobial resistance [8]. The ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) frequently employ biofilm formation as a key resistance mechanism, enabling them to evade conventional antibiotic treatments [8]. Within this context, understanding the nanomechanical properties of matrix polysaccharides through techniques like AFM provides crucial insights for developing novel anti-biofilm strategies.
Polysaccharide Profiles: Structure, Function, and Nanomechanics
Comprehensive Polysaccharide Characteristics
Table 1: Comparative analysis of key biofilm polysaccharides
| Polysaccharide | Primary Organisms | Chemical Structure | Function in Biofilm | AFM Nanomechanical Insights |
|---|---|---|---|---|
| Alginate | Pseudomonas aeruginosa (mucoid variants) | Acetylated copolymer of β-1,4-linked L-guluronic and D-mannuronic acids [9] | Protection from host defenses (scavenges reactive oxygen species), antibiotic resistance, structural integrity [9] | Overproduction creates highly structured, sticky biofilms; architectural changes increase resistance to antimicrobials [10] [9] |
| Psl | Pseudomonas aeruginosa (nonmucoid strains) | Mannose- and galactose-rich polysaccharide [9] | Initial surface attachment, structural scaffold, cell-cell interactions, biofilm architecture maintenance [9] | Serves as receptor for biofilm-tropic bacteriophages [11]; helical surface pattern on individual bacteria [11] |
| Pel | Pseudomonas aeruginosa | Positively charged, partially deacetylated α-1,4-linked N-acetylgalactosamine polymer [12] | Cell-to-cell interactions, cationic matrix component, binds eDNA, aminoglycoside antibiotic protection [12] | Glycoside hydrolase processing alters biofilm biomechanics; exists in cell-associated (HMW) and secreted (LMW) forms with distinct mechanical properties [12] |
| PNAG | Staphylococcus aureus, Escherichia coli and other species | β-1,6-linked N-acetylglucosamine polymer | Adhesion, structural integrity, immune evasion | AFM nanomechanics studies correlate abundance with surface parameters and viability [13] |
Pathophysiological Significance
The clinical implications of these polysaccharides are substantial. Alginate overproduction in P. aeruginosa is particularly associated with worsening clinical prognosis in cystic fibrosis patients, where mucoid variants emerge as the predominant lung pathogen [9]. Alginate appears to protect bacteria from the consequences of inflammation by scavenging free radicals released by activated macrophages and providing protection from phagocytic clearance [9]. Despite antibodies to alginate being found in chronically infected CF patients, these antibodies fail to mediate opsonic killing of P. aeruginosa [9].
Psl plays a critical role in the virulence of P. aeruginosa during infections. It interferes with complement deposition and neutrophil functions, including phagocytosis and ROS production [11]. Moreover, Psl enhances the intracellular survival of phagocytosed P. aeruginosa and improves bacterial survival in mouse models of lung and wound infection [11].
Pel contributes significantly to biofilm resilience through its unique cationic nature, which enables it to bind extracellular DNA (eDNA) and host-derived anionic polymers found in cystic fibrosis sputum, helping to form a structural core and reduce susceptibility to antibiotic and mucolytic treatments [12]. Recent research has demonstrated that glycoside hydrolase processing of the Pel polysaccharide significantly alters biofilm biomechanics and P. aeruginosa virulence in infection models [12].
AFM Nanomechanics in Biofilm Polysaccharide Research
Fundamental AFM Methodologies
Atomic force microscopy provides unparalleled capabilities for characterizing the nanomechanical properties of biofilm polysaccharides under near-physiological conditions. AFM operates by measuring the forces between a sharp tip (typically with radius of 1-20 nm) and the sample surface, with forces in the range of 10⁻¹¹ N to 10⁻⁷ N enabling ultra-sensitive measurements without excessive sample disruption [14]. Key operational modes for biofilm research include:
- Contact Mode: Provides high resolution but generates lateral forces that may be problematic for soft polysaccharide samples [14].
- Tapping Mode: The cantilever oscillates at its resonance frequency, briefly tapping the surface, which minimizes lateral forces and makes it ideal for soft, irregular biofilm samples [14].
- Non-contact Mode: The cantilever vibrates with smaller amplitude without touching the surface, exerting minimal force but offering less precise height measurements [14].
- Force Spectroscopy: Measures force-distance curves to determine interaction forces, adhesion properties, and elastic modulus (Young's modulus) of matrix components [14].
- Phase Imaging: Records the phase shift between the driven cantilever oscillation and its actual response, providing qualitative information about sample hardness, elasticity, and adhesion [14].
Experimental Workflow for Polysaccharide Characterization
The following diagram illustrates a typical AFM workflow for analyzing the nanomechanical properties of biofilm polysaccharides:
Representative Experimental Protocols
AFM Analysis of Alginate Overproduction Effects
Objective: To quantify the effect of alginate overproduction on the nanomechanical properties of P. aeruginosa biofilms [10].
Methodology:
- Cultivate wild-type and mucoid (rpoN mutant) P. aeruginosa strains to establish biofilms
- Use confocal laser scanning microscopy to confirm structural differences in biofilms
- Perform AFM measurements in tapping mode to minimize sample damage
- Collect force-distance curves at multiple locations (minimum 10 points per sample)
- Calculate adhesion forces from retraction curves
- Compare topographic features and stickiness between strains at attachment, establishment, and maturation stages
Key Findings: Biofilms formed by the alginate-overproducing rpoN mutant were significantly stickier during attachment and establishment stages compared to wild-type strains. This difference in stickiness was greatly reduced during the maturation stage, possibly due to cytosolic contents released from dead cells in wild-type biofilms [10].
Nanomechanical Assessment of Capsular Polysaccharides
Objective: To determine the relationship between capsular polysaccharide organization and biofilm formation using AFM nanomechanics [15].
Methodology:
- Prepare isogenic wild-type and capsular mutants of Klebsiella pneumoniae
- Perform in situ nanomechanical measurements using AFM force spectroscopy
- Conduct theoretical modeling of the mechanical data
- Correlate nanomechanical properties with static microtiter plate biofilm assays
- Analyze how type 3 fimbriae influence capsular organization and mechanics
Key Findings: The organization of the capsule significantly influences bacterial adhesion and subsequent biofilm formation. The presence of type 3 fimbriae affects capsular organization, demonstrating the interplay between different surface structures in determining nanomechanical properties relevant to biofilm development [15].
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 2: Key research reagents and materials for polysaccharide and AFM studies
| Reagent/Material | Specifications | Research Application |
|---|---|---|
| Atomic Force Microscope | Equipped with tapping mode, force spectroscopy, and liquid cell capabilities | Nanomechanical characterization of biofilm matrix components under physiological conditions [14] |
| AFM Cantilevers | Si or Si₃N₄ tips with spring constants 0.1-5 N/m, tip radius 1-20 nm | Optimized for soft biological samples; smaller radii provide higher resolution [14] |
| Alginate Lyase | Enzyme specific for alginate degradation | Experimental degradation of alginate to study its functional contribution to biofilm resistance [9] |
| Crystal Violet | 0.1-1% aqueous solution | Standard biofilm biomass quantification through microtiter plate assays [12] |
| PelA Hydrolase Domain (PelAhPa) | Recombinant protein, residues 47-303 | Exogenous addition to disrupt Pel-dependent biofilms; study of Pel polysaccharide processing [12] |
| Conductive AFM Tips | Metal-coated cantilevers with controlled conductivity | Investigation of electrostatic properties of polysaccharides in KPFM mode [14] |
| Psl-Specific Bacteriophages | Clew-1 phage from Bruynoghevirus family | Tool for specifically targeting Psl-containing biofilms; studies of Psl distribution and function [11] |
Biosynthesis Pathways and Regulatory Networks
The biosynthesis of biofilm polysaccharides involves complex pathways with sophisticated regulation. The following diagram illustrates the key biosynthetic and regulatory elements for P. aeruginosa polysaccharides:
Emerging Research Applications and Therapeutic Approaches
Recent advances in understanding polysaccharide nanomechanics have enabled novel therapeutic approaches. One promising strategy involves exploiting the Psl polysaccharide as a receptor for biofilm-tropic bacteriophages. The Clew-1 bacteriophage specifically binds to Psl, enabling it to disrupt P. aeruginosa biofilms and replicate within biofilm bacteria despite inefficiently infecting planktonic cells [11]. This Psl-dependent phage demonstrates reduced bacterial burden in mouse models of P. aeruginosa keratitis, suggesting a targeted approach for biofilm-associated infections [11].
Another innovative approach involves using capsular polysaccharides with antibiofilm activity. Screening of 31 purified capsular polysaccharides revealed that active antibiofilm polymers share distinct biophysical and electrokinetic properties, including high intrinsic viscosity and specific charge characteristics [16]. These non-biocidal polysaccharides, such as Vi, MenA, and MenC, prevent bacterial adhesion without killing cells, potentially reducing selective pressure for resistance [16].
Glycoside hydrolase enzymes present another promising therapeutic avenue. Research has demonstrated that the glycoside hydrolase activity of PelA decreases adherent biofilm biomass and generates the low molecular weight secreted form of Pel exopolysaccharide, which in turn influences biofilm biomechanics and reduces P. aeruginosa virulence in Caenorhabditis elegans and Drosophila melanogaster infection models [12]. This suggests that engineered hydrolases could be developed to modulate biofilm properties and enhance susceptibility to conventional antibiotics.
The integration of AFM nanomechanics with biofilm research has provided unprecedented insights into the structure-function relationships of key polysaccharides. Alginate, Psl, Pel, and PNAG each contribute distinct mechanical and protective properties to the biofilm matrix, enabling bacterial communities to withstand environmental stresses, antimicrobial treatments, and host immune responses. The continuing refinement of AFM methodologies, combined with emerging therapeutic approaches that target these polysaccharides, holds significant promise for combating biofilm-associated infections. Future research should focus on correlating nanomechanical measurements with clinical outcomes to facilitate the translation of these findings into effective anti-biofilm strategies.
The Capsule as a Critical Virulence Determinant
The bacterial capsule is a dense, well-structured polymer layer that surrounds the cell envelope of many bacterial pathogens. Primarily composed of high molecular weight polysaccharides (capsular polysaccharides, or CPS), this layer forms a viscous, hydrated gel that constitutes the outermost interface between the pathogen and its host environment [17]. In Gram-negative bacteria, these polymers are often covalently anchored to lipids in the outer membrane, while in Gram-positive bacteria, they are typically attached to the peptidoglycan cell wall [18]. Historically, the capsule was first described as a "halo" around bacterial cells by Pasteur in 1881, with its carbohydrate nature elucidated by Avery in 1925 [17]. Far from being a mere structural component, the capsule is now recognized as a critical virulence factor that enables pathogens to evade host immune defenses, establish infections, and persist in hostile environments. Its role is particularly crucial in biofilm formation, where it contributes to initial adhesion, structural stability, and protection against antimicrobial agents. The following sections provide an in-depth examination of the capsule's biosynthesis, its multifaceted functions in virulence, and the advanced nanomechanical techniques used to study its properties, with a specific focus on insights gained from atomic force microscopy (AFM) in biofilm research.
Biosynthesis and Structural Diversity of Capsules
Biosynthetic Pathways
Bacteria synthesize their capsular polysaccharides through several distinct biochemical pathways, with the choice of pathway depending on the bacterial species and the specific capsule type. The most prevalent mechanisms are the Wzx/Wzy-dependent pathway, the ATP-binding cassette (ABC) transporter-dependent pathway, and the synthase-dependent pathway [17].
The Wzx/Wzy-dependent pathway is employed by over 90% of Streptococcus pneumoniae serotypes and is fundamental to Group I and IV capsules in Gram-negative bacteria [17]. This mechanism involves the assembly of oligosaccharide repeat units on a lipid carrier (undecyl isoprene phosphate) on the cytoplasmic face of the inner membrane. The Wzx "flippase" enzyme then translocates these units across the membrane, and the Wzy polymerase links them together to form the full-length polysaccharide chain [17]. In Klebsiella pneumoniae, a leading model organism for capsule studies, this process is initiated by WbaP, a galactose phosphotransferase that links galactose to the undecaprenyl phosphate lipid carrier [19]. Subsequent glycosyltransferases add further sugars, with the Wzx flippase moving the growing chain to the periplasm where Wzy polymerase extends it. The tyrosine kinase Wzc regulates chain length, and the outer membrane protein Wza exports the completed polysaccharide, which is then anchored to the cell surface by Wzi [19].
In contrast, the ABC transporter-dependent pathway involves complete polymerization of the polysaccharide chain in the cytoplasm before it is transported across the inner membrane by an ABC transporter complex [17]. While the Wzx/Wzy and ABC transporter mechanisms differ in their assembly sites, both utilize outer membrane proteins from the polysaccharide export family to transport the finished capsule across the outer membrane of Gram-negative bacteria.
Less common is the synthase-dependent mechanism, utilized by S. pneumoniae serotypes 3 and 37, where a single enzyme complex is responsible for both polymerization and translocation of the capsule [17]. Some bacteria, notably Bacillus anthracis, produce capsules composed primarily of polypeptides (poly-D-glutamic acid) rather than polysaccharides, with biosynthesis involving the transfer of activated glutamate units to a membrane-bound polyglutamyl acceptor chain [17].
Capsular Serotypes and Virulence
Capsular types are distinguished by variations in monosaccharide composition, glycosidic linkage positions, stereochemistry (L or D configuration), and chemical modifications such as O-acetylation [17]. This structural diversity gives rise to numerous serotypes (or serovars), which are critical for classifying bacterial strains and understanding their pathogenic potential.
- Escherichia coli produces approximately 80 distinct capsule types, categorized into four groups (I-IV), including the polysialic acid (PSA)-containing K1 capsule and the heparosan-containing K5 capsule [17].
- Streptococcus pneumoniae has 93 identified capsular serotypes, with significant variation in their invasive potential. Serotypes 1, 4, 5, 8, 12F, 18C, and 19A are highly invasive, while 6A, 6B, 11A, and 23F are less aggressive [17].
- Klebsiella pneumoniae produces 77 recognized CPS serotypes, with the K2 serotype being particularly associated with bacteremia and hypervirulence [19].
The virulence of different serotypes is closely linked to their specific chemical structures. For instance, highly virulent K. pneumoniae K2 strains lack the mannose-α-2/3-mannose structure present in less virulent strains [17]. Furthermore, pathogens can undergo capsular switching through genetic recombination, altering their surface antigens to evade host immunity. For example, S. pneumoniae serotype 11E evolves from serotype 11A through mutations in the wcjE gene, enabling escape from ficolin-2-mediated phagocytosis during invasive disease [17]. This structural plasticity underscores the capsule's role in adaptive virulence and presents a challenge for both typing and therapeutic targeting. Advanced computational tools like Kaptive Web have been developed to rapidly type Klebsiella K and O loci, facilitating the identification of capsule types associated with high virulence [17].
The Capsule as a Virulence Factor
The capsule contributes to bacterial pathogenicity through multiple mechanisms, primarily by acting as a physical barrier that impedes host immune recognition and response. Its functions span from initial adhesion to survival within the host, making it indispensable for successful infection.
Immune Evasion and Serum Resistance
The capsule's most critical role is protecting bacteria from phagocytosis by host immune cells. The polysaccharide layer masks antigenic surface proteins, preventing recognition by phagocytic receptors [19]. This protective effect is clearly demonstrated in Klebsiella pneumoniae, where the capsule is essential for survival in human blood. Wild-type encapsulated K. pneumoniae can resist phagocytosis and complement-mediated killing, whereas non-encapsulated mutants (e.g., Kpn2146Δwza) are rapidly cleared in whole blood and plasma assays [19]. In vivo experiments using Galleria mellonella larvae infection models confirm the dramatically decreased virulence of capsule-deficient mutants [19].
A particularly effective immune evasion strategy involves the production of capsules that mimic host tissue molecules. Capsules composed of polysialic acid (PSA), hyaluronan (HA), heparosan, or chondroitin are structurally identical to mammalian glycans [18]. This molecular mimicry renders these capsules "self" antigens, preventing the host from mounting an effective antibody response. Pathogens bearing such non-immunogenic coatings, including E. coli K1 (PSA) and Group A Streptococcus (HA), can thus reside in host tissues for extended periods, reaching high population densities that enable disease progression [18].
Adhesion, Invasion, and Biofilm Formation
While the capsule was traditionally thought to inhibit adhesion by sterically blocking adhesins, research reveals a more nuanced relationship. The capsule plays a context-dependent role in bacterial interactions with host cells. In K. pneumoniae, for instance, encapsulated and non-encapsulated strains show similar adherence levels to A549 lung epithelial cells, but the non-encapsulated mutant exhibits moderately higher internalization, suggesting the capsule may limit invasion into certain cell types [19].
In biofilm formation, the capsule and other exopolysaccharides are fundamental components of the extracellular polymeric substance (EPS) matrix [20]. Biofilm development proceeds through defined stages: (1) reversible attachment, (2) irreversible attachment, (3) microcolony formation, (4) maturation, and (5) dispersion [21] [22]. The capsule contributes to multiple stages of this process. During initial attachment, the capsule can facilitate bacterial approach to surfaces through non-specific interactions. During irreversible attachment and maturation, the capsule provides structural integrity and stability to the biofilm architecture [15].
The capsule's organization and properties are influenced by other surface structures, particularly type 3 fimbriae, which can affect capsular organization and thereby modulate bacterial adhesion and biofilm development [15]. In Pseudomonas aeruginosa, a model organism for biofilm studies, exopolysaccharides like Psl, Pel, and alginate are crucial for surface attachment, structural stability, and resistance to environmental stresses [21]. The secondary messenger cyclic di-GMP (c-di-GMP) is a central regulator of this process; high intracellular levels of c-di-GMP promote exopolysaccharide production and repress flagellar motility, facilitating the transition from planktonic to biofilm growth [21] [22].
Table 1: Key Virulence Functions of Bacterial Capsules
| Function | Mechanism | Example Pathogens |
|---|---|---|
| Phagocytosis Resistance | Masks surface antigens; prevents complement deposition and opsonization | Klebsiella pneumoniae, Streptococcus pneumoniae [19] |
| Molecular Mimicry | Composed of polysaccharides identical to host glycans (e.g., HA, PSA) | E. coli K1 (PSA), Group A Streptococcus (HA) [18] |
| Biofilm Formation | Contributes to EPS matrix; aids in adhesion and structural stability | Pseudomonas aeruginosa, Klebsiella pneumoniae [15] [20] |
| Environmental Adaptation | Acts as a hydrogel to protect against osmotic stress and desiccation | Klebsiella pneumoniae [23] |
| Antibiotic Resistance | Limits antimicrobial penetration through the biofilm matrix | ESKAPE pathogens (K. pneumoniae, P. aeruginosa, etc.) [20] |
AFM Nanomechanics in Capsule and Biofilm Research
Atomic force microscopy has emerged as a powerful tool for investigating the nanomechanical properties of bacterial capsules and their role in biofilm formation. Unlike traditional microscopic techniques, AFM allows for in situ quantitative measurements of living bacterial cells under physiological conditions, providing unprecedented insights into the biophysical behavior of the capsular layer.
Probing Capsular Mechanics and Organization
AFM-based force spectroscopy involves bringing a functionalized tip into contact with the bacterial surface and retracting it to obtain force-distance curves. These curves reveal information about the elasticity, adhesion, and topography of the capsule at the nanoscale.
Seminal AFM studies on Klebsiella pneumoniae have demonstrated that the polysaccharide capsule behaves as a responsive polymer hydrogel [23]. This hydrogel structure can undergo rapid, reversible collapse and recovery in response to changes in osmotic pressure, acting as an "ion sponge" that protects the cell from osmotic stress [23]. This adaptive property is crucial for survival in diverse host environments.
Furthermore, AFM has elucidated the relationship between capsule organization and biofilm formation. Theoretical modeling of AFM nanomechanical data shows that the spatial organization of the capsule significantly influences bacterial adhesion—the critical first step in biofilm development [15] [24]. The presence of surface appendages like type 3 fimbriae can alter capsular organization, thereby modulating the bacterium's adhesive properties and its potential to form biofilms [15].
Experimental Workflow for AFM Analysis
A typical AFM nanomechanics experiment targeting bacterial capsules involves the following key steps:
- Strain Selection and Mutant Generation: Isogenic mutants deficient in capsule synthesis (e.g., Δwza mutants lacking the exporter protein) or other related structures (e.g., fimbriae) are constructed for comparison with the wild-type strain [19] [15]. This allows for direct attribution of observed effects to the capsule.
- Sample Preparation: Bacterial cells are cultured under appropriate conditions to ensure capsule expression. Cells are then immobilized onto a solid substrate (e.g., a poly-L-lysine-coated glass slide or membrane filter) without chemical fixation to preserve native capsule structure and mechanical properties [15] [24].
- AFM Force Measurement: A cantilever with a defined tip (often silicon nitride) is used to probe the surface of immobilized cells in a liquid environment. Hundreds of force-distance curves are acquired across the surface of multiple individual bacteria to ensure statistical robustness [15] [23].
- Data Analysis: Force curves are analyzed to extract nanomechanical parameters, including:
- Young's Modulus: A measure of cellular stiffness, indicating the deformability of the capsule.
- Adhesion Force: The force required to separate the tip from the surface, reflecting the stickiness of the capsular layer.
- Turgor Pressure: The internal pressure of the cell, which can be inferred from the force curves [23].
- Correlation with Phenotypic Assays: AFM data is correlated with results from standard microbiological assays, such as static microtiter plate biofilm assays, to link nanomechanical properties with macroscopic phenotypic outcomes like biofilm formation capacity [15].
Diagram 1: AFM Nanomechanics Workflow for Capsule Study. This flowchart outlines the key experimental steps, from biological preparation to data interpretation, for investigating bacterial capsules using Atomic Force Microscopy.
Experimental Protocols and Research Toolkit
This section provides detailed methodologies for key experiments used to elucidate the role of the capsule in virulence and biofilm formation, serving as a practical guide for researchers.
Protocol 1: Generation of a Capsule-Deficient Mutant
The construction of an isogenic, capsule-deficient mutant is a fundamental step for comparative studies. The following protocol, adapted from a study on Klebsiella pneumoniae, targets the wza gene, which encodes a conserved outer membrane polysaccharide export protein [19].
- Primer Design: Design primers to amplify the target gene (wza) along with approximately 500 base pairs of its upstream and downstream flanking regions. This ensures homologous recombination.
- Cloning:
- Amplify the wza region with flanking sequences via PCR and ligate the product into a suitable cloning vector (e.g., pMiniT).
- Transform the construct into an E. coli host strain (e.g., 10β) and select for transformants.
- Subclone the insert into a suicide vector (e.g., pKOV) that cannot replicate in the target pathogen.
- Allelic Exchange:
- Perform an inverse PCR on the suicide vector to remove the wza coding sequence.
- Insert a selectable marker (e.g., a spectinomycin resistance gene, aad9) in place of the deleted wza gene.
- Transform the final suicide vector (pKOV-∆wza_specR) into the wild-type K. pneumoniae strain via conjugation or electroporation.
- Select for clones where a double-crossover event has replaced the wild-type wza allele with the mutant construct, resulting in a clean, non-polar deletion. Confirm the mutation via PCR and sequencing [19].
Protocol 2: AFM Nanomechanics of Bacterial Cells
This protocol details the procedure for measuring the nanomechanical properties of encapsulated bacteria [15] [24] [23].
Bacterial Culture and Immobilization:
- Grow the wild-type and isogenic capsule mutant strains to mid-log phase under conditions that promote capsule expression.
- Gently harvest cells by centrifugation to avoid damaging the capsule. Wash and resuspend in an appropriate buffer (e.g., PBS or a defined minimal medium).
- Immobilize cells by depositing a droplet of the bacterial suspension onto a poly-L-lysine-coated glass slide or a membrane filter for 15-30 minutes. Rinse carefully with buffer to remove non-adhered cells.
AFM Force Spectroscopy:
- Mount the sample on the AFM liquid cell and immerse in the same buffer.
- Use a sharp, non-functionalized silicon nitride cantilever with a known spring constant (calibrated prior to measurements).
- Approach the central surface of a single, well-isolated bacterial cell and collect force-distance curves using a trigger force of 250-500 pN to avoid damaging the cell.
- Collect a grid of at least 256 force curves (16x16) on each cell and measure a minimum of 10 different cells per strain.
Data Processing and Analysis:
- Use appropriate software (e.g., the AFM manufacturer's software, Igor Pro, or custom scripts) to analyze the force curves.
- Fit the extending part of the force curve with a suitable model (e.g., Hertzian, Sneddon, or Johnson-Kendall-Roberts models) to derive the Young's modulus, a measure of cell stiffness.
- Analyze the retraction curves to quantify adhesion forces and work of adhesion.
- Statistically compare the distributions of Young's modulus and adhesion force between wild-type and mutant strains to determine the mechanical contribution of the capsule.
Table 2: Research Reagent Solutions for Capsule and Biofilm Studies
| Reagent / Material | Function / Application | Example Use Case |
|---|---|---|
| Suicide Vector (e.g., pKOV) | Facilitates allelic exchange for generating knockout mutants | Construction of isogenic capsule-deficient mutant (∆wza) in K. pneumoniae [19] |
| Poly-L-Lysine Coated Substrates | Promotes electrostatic immobilization of bacterial cells for AFM | Securing live Klebsiella cells for nanomechanical measurements without chemical fixation [15] |
| Silicon Nitride AFM Cantilevers | Probes for nanomechanical force spectroscopy; can be functionalized | Measuring elasticity and adhesion of the bacterial capsule in liquid [15] [23] |
| c-di-GMP Assay Kits | Quantify intracellular cyclic di-GMP levels, a key regulator of EPS production | Linking second messenger signaling to capsule production and biofilm phenotype [21] |
| Specific Glycosidases / Depolymerases | Enzymatically digest specific capsule polysaccharides | Confirming capsule composition; studying dispersal of capsule-dependent biofilms [17] |
Therapeutic Implications and Future Directions
Understanding the capsule's role in virulence and its nanomechanical properties opens promising avenues for novel anti-infective strategies. The primary therapeutic approaches include:
- Capsule-Targeting Vaccines: Vaccines based on purified capsular polysaccharides have been successful against pathogens like S. pneumoniae and H. influenzae type b (Hib) [17]. Conjugating CPS to carrier proteins converts the T-cell-independent immune response to a T-cell-dependent one, improving immunogenicity and long-lasting immunity, especially in children [18].
- Enzymatic Dispersal: Enzymes that degrade the capsule or biofilm matrix represent a powerful therapeutic approach. For example, dispersin B degrades PNAG (poly-N-acetylglucosamine), a biofilm polysaccharide produced by staphylococci and E. coli [20]. Similarly, bacteriophage-derived depolymerases that specifically cleave capsule polysaccharides can sensitize bacteria to antibiotic treatment and host immune clearance [17].
- Inhibition of Biosynthesis and Regulation: Small molecules that disrupt capsule biosynthesis pathways or inhibit regulatory systems (like the c-di-GMP signaling network) could prevent capsule production, rendering the pathogen susceptible to host defenses [21]. The Wzx/Wzy pathway is a particularly attractive target for such inhibitors.
- Nanotechnology-Based Delivery: Nanoparticles can be engineered to deliver antibiotics or biofilm-disrupting agents directly to the site of infection, potentially penetrating the capsular barrier more effectively than free drug molecules.
The application of AFM and other biophysical techniques continues to refine our understanding of the capsule as a dynamic, responsive structure. Future research will likely focus on mapping the spatial heterogeneity of capsules within biofilms and investigating the real-time structural changes during infection. This knowledge, integrated with genomics and immunology, will be crucial for designing the next generation of antimicrobial therapies that effectively neutralize this critical virulence determinant.
Diagram 2: Therapeutic Strategies Targeting the Capsule. This diagram illustrates how different therapeutic approaches (ellipses) interact with specific biological targets (boxes) associated with the bacterial capsule and its functions.
Linking Capsular Organization to Bacterial Adhesion and Aggregation
In the field of bacterial pathogenesis, the bacterial capsule—a surface-associated polymer—and its role in adhesion present a seeming paradox. While capsules are recognized virulence factors that can resist phagocytosis, their presence might intuitively be expected to hinder the initial adhesion to surfaces, a critical first step in biofilm formation and infection [25] [17]. This review delves into the resolution of this paradox, exploring how the specific structural organization of the capsule, rather than its mere presence or absence, dictates bacterial adhesion and aggregation behaviors. Framed within the context of atomic force microscopy (AFM) nanomechanics, this guide synthesizes current research to elucidate the biophysical mechanisms through which capsules modulate bacterial interactions with surfaces and other cells, ultimately influencing biofilm architecture and virulence.
Capsular Polysaccharides: Structure, Biosynthesis, and Function
Diversity and Biosynthesis
Bacterial capsules are polymers, primarily consisting of high-molecular-weight polysaccharides, secreted at the periphery of the cell wall and enveloping the entire cell [17]. In Escherichia coli, approximately 80 distinct capsular polysaccharides (K antigens) have been identified, categorized into four major groups (I-IV) [25] [17]. These include structurally diverse polymers such as polysialic acid (K1), chondroitin (K4), and heparosan (K5) [17]. Three primary pathways for capsule synthesis are recognized: the Wzx/Wzy-dependent mechanism, the ATP-binding cassette (ABC) transporter-dependent mechanism, and the synthase-dependent mechanism [17]. The Wzx/Wzy-dependent pathway, used by over 90% of Streptococcus pneumoniae serotypes, involves the formation of repeat units on a lipid carrier, which are then "flipped" across the membrane by the Wzx flippase and polymerized by Wzy [17].
Biological Roles in Virulence
The capsule is a quintessential virulence determinant. Its functions extend beyond adhesion modulation to include:
- Resistance to Phagocytosis: Capsules physically shield bacterial surface components from recognition by immune cells and can be anti-phagocytic [25] [17].
- Serum Resistance: Certain capsules, such as the K1 capsule in E. coli, confer resistance to the bactericidal action of human serum, enhancing survival in blood and tissues [25].
- Protection from Desiccation and Antimicrobials: The capsule provides a protective barrier against environmental stresses and some antimicrobial agents [25] [17].
Table 1: Primary Capsule Biosynthesis Pathways in Bacteria
| Pathway | Key Features | Representative Bacteria/Serotypes |
|---|---|---|
| Wzx/Wzy-dependent | Repeat units formed on lipid carrier; flipped by Wzx and polymerized by Wzy [17]. | >90% of S. pneumoniae serotypes [17]; E. coli Group I & IV [17]. |
| ABC Transporter-dependent | Polysaccharide chains polymerized in cytoplasm and transported by ABC transporters [17]. | E. coli Group II & III [17]. |
| Synthase-dependent | A single synthase enzyme initiates, polymerizes, and translocates the capsule [17]. | S. pneumoniae serotypes 3 & 37 [17]. |
The Adhesion-Aggregation Paradox: Steric Shielding vs. Facilitation
A central question in bacterial surface interactions is how a substantial capsular layer, which can extend 0.2 to 1.0 μm from the cell surface, affects the function of shorter bacterial adhesins, such as Antigen 43 (Ag43) and AIDA-I, which protrude only about 10 nm [25]. Research has demonstrated that the capsule can sterically shield these shorter adhesins, physically blocking their receptor-target recognition and thereby abolishing Ag43-mediated cell-to-cell aggregation [25]. This phenomenon is not restricted to E. coli but can occur in other Gram-negative bacteria [25].
However, this negative interference is not absolute. The organizational state of the capsule is critical. A compact or organized capsule may indeed block adhesion, while a disorganized or loosely bound capsule may allow adhesins to function. This organizational change can be influenced by other surface structures, such as type 3 fimbriae, which can rearrange the capsule, potentially creating microdomains where adhesins can effectively engage with their targets [15]. Furthermore, in some contexts, the capsule itself can act as an adhesive, facilitating a more generalized, weak interaction with surfaces that precedes the stronger, specific interactions mediated by adhesins [15].
AFM Nanomechanics: Probing Bacterial Surfaces and Adhesive Forces
Atomic Force Microscopy (AFM) is a high-resolution form of scanning probe microscopy that has become an indispensable tool for quantifying the nanomechanical properties of bacterial surfaces [26]. The technique operates by scanning a sharp tip on the end of a cantilever over the sample surface. As the tip interacts with the sample surface, attractive or repulsive forces cause cantilever deflection, which is measured by a laser reflected into photodiodes [26]. A scanner controls the probe height, and the variance in height is used to produce a three-dimensional topographical representation [26].
Key AFM Modologies in Biofilm Research
- Contact Mode: The tip maintains constant contact with the sample surface, providing constant force and fast scanning times, though it may deform soft samples [26].
- Tapping Mode: The cantilever is oscillated at its resonance frequency, and the tip gently "taps" the surface. This mode is gentler and provides higher lateral resolution for soft, adhesive samples like bacterial cells, minimizing sample damage [26].
- Noncontact Mode: The cantilever oscillates just above its resonance frequency without touching the sample, exerting minimal force. However, it typically offers lower resolution [26].
AFM's application in biofilm research allows for the in situ measurement of bacterial adhesion forces. By functionalizing the AFM tip with specific molecules (e.g., adhesins, host receptors) or using a single bacterial cell as a probe, researchers can directly quantify the nanomechanical forces governing capsule deformation, cell-surface adhesion, and cell-cell aggregation [15].
Figure 1: AFM Experimental Workflow for Bacterial Capsule Study. This diagram outlines the key steps in an AFM-based nanomechanics study, from mode selection to data analysis.
Quantitative AFM Findings on Capsular Organization and Adhesion
AFM nanomechanics studies have provided direct, quantitative evidence linking capsular organization to adhesion. A seminal study on Klebsiella pneumoniae compared wild-type strains with specific mutants using AFM and found that the organization of the capsule, influenced by the presence of type 3 fimbriae, directly affected bacterial adhesion and biofilm formation [15]. Theoretical modeling of the mechanical data from these experiments supported the conclusion that a structured capsule can hinder adhesion, while its reorganization can facilitate it [15].
The steric blocking effect has been quantitatively demonstrated for short adhesins like Antigen 43 (Ag43). In encapsulated E. coli strains, the presence of a capsule was shown to block Ag43-mediated aggregation, a function that was restored upon loss of the capsule [25]. This supports the model where the capsule acts as a physical barrier, preventing the close cell-to-cell contact (typically within 10-12 nm) required for short adhesins to engage in homophilic or heterophilic binding [25].
Table 2: Key Quantitative Findings from AFM and Related Studies on Capsular Function
| Bacterial System | Experimental Manipulation | Key Quantitative Finding | Impact on Adhesion/Aggregation |
|---|---|---|---|
| E. coli [25] | Presence vs. absence of capsule | Capsule blocks Ag43 function (≈10 nm protrusion) via steric shielding [25]. | Abolished cell-to-cell aggregation; reduced biofilm formation [25]. |
| Klebsiella pneumoniae [15] | Wild-type vs. fimbriae-deficient mutants | Fimbriae alter capsular organization, measured via AFM nanomechanics [15]. | Significant change in adhesion force; modulated biofilm formation [15]. |
| Various Gram-negative bacteria [25] | Heterologous expression of Ag43 | Ag43-mediated aggregation is a general phenomenon that is disrupted by capsule [25]. | Cell aggregation is consistently blocked in the presence of a capsule [25]. |
Detailed Experimental Protocol: AFM-Based Analysis of Bacterial Adhesion
The following protocol outlines the key steps for assessing the role of capsular organization in bacterial adhesion using AFM, synthesizing methodologies from cited studies.
Bacterial Strain Selection and Growth
- Strains: Utilize isogenic pairs of wild-type and mutant strains (e.g., capsule-deficient mutants, fimbriae-deficient mutants) [25] [15]. For example, E. coli strains with and without the K1 or K5 capsule operon [25].
- Growth Conditions: Grow bacteria in appropriate liquid media (e.g., Luria-Bertani broth) at 37°C to mid-exponential phase (OD600 ≈ 0.5-0.8) to ensure consistent capsule expression [25]. The culture conditions should be meticulously controlled and reported, as capsule expression can be influenced by growth phase and medium composition.
Sample Preparation for AFM
- Immobilization: Gently immobilize bacterial cells onto a solid substrate to prevent detachment during scanning. This can be achieved by:
- Poly-L-lysine Coating: Deposit a drop of bacterial suspension onto a freshly cleaved mica surface pre-coated with 0.1% (w/v) poly-L-lysine for 10-15 minutes [26].
- Mild Chemical Fixation: Optionally, use a low concentration of glutaraldehyde (e.g., 0.5-1.0%) for a short duration (5-10 min) to stabilize the cells without significantly altering surface mechanics, though this may affect native protein function.
- Washing: Gently rinse the sample with a suitable buffer (e.g., phosphate-buffered saline) to remove non-adherent cells and media components. The sample should be kept hydrated at all times.
AFM Imaging and Force Spectroscopy
- Instrumentation: Use a commercial AFM system (e.g., Bruker Multimode) [27].
- Probes: Select sharp, cantilevers with a nominal spring constant of ~0.01-0.1 N/m for force spectroscopy on soft biological samples. The probe type and approximate resonant frequency (e.g., AppNano ACT probes, ~300 kHz) should be specified [27].
- Imaging Mode: Tapping mode is highly recommended for initial topographical imaging to minimize sample damage and obtain high-resolution images of the soft, hydrated bacterial surface [26].
- Force Measurement:
- Approach-Retract Cycles: Program the AFM to perform multiple force-distance curves (≥ 1000 per sample) on different locations of the cell surface.
- Adhesion Force Quantification: The retraction curve often shows a characteristic "pull-off" event, the magnitude of which corresponds to the unbinding force or adhesion force between the tip and the sample surface.
- Tip Functionalization (Optional): To measure specific interactions (e.g., adhesin-receptor binding), the AFM tip can be functionalized with relevant proteins or sugars using linkers like PEG-bis(N-succinimidyl succinate).
Data Analysis and Statistical Validation
- Adhesion Force and Probability: Calculate the average adhesion force and the adhesion probability (percentage of force curves showing adhesion events) from the collected force-distance curves.
- Statistical Testing: Perform appropriate statistical tests (e.g., Student's t-test for comparing two groups) to determine if differences in adhesion forces between strains are significant. Data should be collected from a minimum of three independent biological replicates.
- Topographical Analysis: Use AFM image analysis software (e.g., Gwyddion) to determine surface roughness and capsule dimensions [27].
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Reagents and Materials for AFM Studies of Bacterial Capsules
| Item Category | Specific Example(s) | Function and Application in Research |
|---|---|---|
| Bacterial Strains | Isogenic wild-type and mutant pairs (e.g., E. coli K-12 with flu (Ag43) mutation, E. coli 1177 (O1:K1) with fim mutation) [25]. | Enable direct comparison of the functional impact of specific genes (e.g., adhesins, capsule biosynthesis, fimbriae) on adhesion and aggregation [25]. |
| Molecular Cloning Plasmids | pKKJ128 (carries flu gene for Ag43 expression), pKT274 (carries K1 capsule gene operon) [25]. | Used for genetic manipulation to complement mutations or heterologously express capsule and adhesin genes in different bacterial backgrounds [25]. |
| AFM Probes | AppNano ACT probes (nominal frequency ~300 kHz) [27]. | The physical probe that interacts with the sample; its stiffness and sharpness determine resolution and force sensitivity. |
| AFM Substrates | Freshly cleaved mica [26]. | Provides an atomically flat, clean surface for immobilizing bacterial cells for high-resolution AFM imaging. |
| Immobilization Reagents | Poly-L-lysine solution (0.1% w/v) [26]. | Promotes electrostatic adhesion of bacterial cells to the mica substrate, preventing them from being displaced by the AFM tip. |
| Software for Analysis | Gwyddion, SPIP [27]. | Used for processing and analyzing AFM images, including measuring surface roughness, particle dimensions, and generating 3D renderings. |
Figure 2: Logical Relationship Between Capsular Organization and Biofilm Outcome. This diagram illustrates the causal pathway from factors influencing capsule organization to the ultimate impact on biofilm formation, as revealed by AFM studies.
Polysaccharides are fundamental components of biological systems, playing a critical role in determining the mechanical stability of structures ranging from bacterial biofilms to marine gels and synthetic lipid vesicles. Within the context of AFM nanomechanics studies on capsular polysaccharides in biofilm research, this review examines the specific biophysical mechanisms through which polysaccharides confer mechanical integrity. We integrate quantitative force spectroscopy data, detailed experimental protocols, and structural models to provide researchers and drug development professionals with a comprehensive technical resource for understanding and investigating polysaccharide-mediated mechanical stabilization, highlighting how these principles can be leveraged for therapeutic intervention and biomaterial design.
The mechanical properties of biological assemblies are frequently dictated by polysaccharide components that provide structural integrity, mediate environmental interactions, and determine functional behavior. In biofilm research, understanding how capsular polysaccharides confer mechanical stability is paramount for developing strategies to combat biofilm-associated infections. Atomic force microscopy (AFM) nanomechanics has emerged as a powerful technique for quantifying these properties at the molecular and cellular levels, revealing intricate relationships between polysaccharide structure and mechanical function [15]. This technical guide synthesizes current understanding of the primary biophysical mechanisms involved, supported by quantitative data and standardized methodologies to enable reproducible research in this evolving field. The insights gained not only elucidate fundamental biological principles but also inform drug delivery system design by revealing how polymer functionalization modulates mechanical properties of synthetic vesicles [28].
Quantitative Data on Polysaccharide Mechanical Properties
The mechanical behavior of polysaccharides has been quantified through various experimental approaches, revealing distinct patterns that correspond to specific structural configurations and interaction modalities. The following tables summarize key quantitative findings from recent studies.
Table 1: Mechanical Signatures of Polysaccharides in Various Systems
| System Studied | Experimental Technique | Mechanical Signature | Quantified Parameters | Structural Interpretation |
|---|---|---|---|---|
| Marine-gel biopolymers [29] | AFM Force Spectroscopy | Force-extension curves with entropic elasticity followed by chair-to-boat transitions | N/A (qualitative patterns) | Individual polysaccharide fibrils undergoing conformational changes |
| Marine-gel biopolymers (Low cross-linking) [29] | AFM Force Spectroscopy | Sawtooth patterns | N/A (qualitative patterns) | Unraveling of polysaccharide entanglements under applied force |
| Marine-gel biopolymers (High cross-linking) [29] | AFM Force Spectroscopy | Force plateaus | N/A (qualitative patterns) | Unzipping and unwinding of helical bundles |
| Marine-gel biopolymers (3D network) [29] | AFM Force Spectroscopy | Force staircases of increasing height | N/A (qualitative patterns) | Hierarchical peeling of fibrils from junction zones |
| Klebsiella pneumoniae biofilms [15] | AFM Nanomechanics | Modified adhesion properties | N/A (qualitative study) | Capsular organization influenced by fimbriae presence |
Table 2: Impact of Chondroitin Sulfate Functionalization on Lipid Vesicle Mechanics
| Vesicle Type | Experimental Technique | Stiffness Metric | Control Value | Modified Value | Change |
|---|---|---|---|---|---|
| DOPC GUVs [28] | Micropipette Aspiration | Stretching Modulus | Reference value | Reduced | Softening effect |
| DOPC GUVs [28] | Fluorescence Recovery After Photobleaching (FRAP) | Diffusion Coefficient | 2.32 ± 0.23 μm²/s | Decreased by >10% | Reduced membrane fluidity |
| Lipid Vesicles with PEG [28] | Micropipette Aspiration | Stretching Modulus | Reference value | Unchanged or increased | No reduction in stiffness |
Table 3: Polysaccharide Coating Density Calculations
| Parameter | Value | Method of Determination |
|---|---|---|
| Chol-CS saturation concentration [28] | 50 nM | Fluorescence intensity analysis of membrane-bound vs. free Chol-CS |
| Estimated membrane density [28] | 0.4 mol% | Calculation from confocal microscopy data |
| Intermolecular distance [28] | ~6.6 nm | Based on DOPC headgroup area (0.7 nm²) |
| Hydrodynamic radius of Chol-CS [28] | >6.6 nm | Dynamic light scattering (referenced in SI) |
Experimental Protocols and Methodologies
AFM Force Spectroscopy for Polysaccharide Networks
Atomic force microscopy force spectroscopy experiments enable the quantification of intra- and intermolecular forces within polysaccharide networks [30]. The standard protocol involves:
Sample Preparation: Marine-gel biopolymers or purified polysaccharides are deposited on freshly cleaved mica substrates and allowed to adsorb for 15-60 minutes in appropriate buffer conditions [29].
Cantilever Selection and Calibration: Soft cantilevers with spring constants of 0.01-0.1 N/m are recommended for polysaccharide studies. The precise spring constant must be determined using thermal tuning methods or reference samples [30].
Force Curve Acquisition: The AFM tip is brought into contact with the sample surface and then retracted at constant velocity (typically 0.5-1 μm/s). Thousands of force-distance curves are collected at multiple locations to build statistical understanding [30] [29].
Data Analysis:
- Convert cantilever deflection to force using Hooke's law (F = -k×d, where k is the spring constant and d is deflection)
- Identify characteristic patterns in retraction curves: entropic elasticity, sawtooth patterns, force plateaus, or force staircases
- Fit polymer stretching models (e.g., Worm-like Chain or Freely Jointed Chain) to quantify persistence lengths and contour lengths
- Map specific mechanical signatures to structural features through correlation with known chemical composition [29]
Micropipette Aspiration for Vesicle Mechanics
The effect of polysaccharides on membrane mechanical properties can be quantified using micropipette aspiration:
GUV Formation: Giant unilamellar vesicles are formed via electroformation in sucrose solution (100-200 mOsm) using lipid compositions of interest [28].
Asymmetric Functionalization: Cholesterol-conjugated polysaccharides (e.g., Chol-CS) are added to pre-formed GUVs at concentrations up to 50 nM saturation point and incubated for 30-60 minutes to allow incorporation into the outer leaflet [28].
Aspiration Setup:
- Transfer GUVs to iso-osmotic glucose solution in observation chamber
- Use micropipettes with diameters of 5-10 μm
- Apply controlled suction pressure (ΔP) of approximately 1 Pa using precision pressure control system [28]
Stretching Modulus Calculation:
- Measure the change in projection length (ΔL) inside the pipette versus applied pressure
- Calculate the stretching modulus (K) using the formula: K = (ΔP × R₀²) / (ΔL × Rp) where R₀ is the initial vesicle radius and Rp is the pipette radius
- Compare functionalized versus control vesicles to quantify polysaccharide-induced softening [28]
AFM Nanomechanics of Bacterial Biofilms
To study the role of capsular polysaccharides in biofilm formation:
Bacterial Strain Preparation: Wild-type and isogenic mutants (e.g., fimbriae-deficient strains) of relevant pathogens such as Klebsiella pneumoniae are cultured under conditions that promote capsule expression [15].
Sample Immobilization: Bacterial cells are chemically immobilized on functionalized glass substrates using poly-L-lysine or specific antibodies to maintain viability while preventing movement during AFM imaging [15].
In Situ Nanomechanical Measurements:
- Perform force mapping over multiple bacterial cells (typically 16×16 or 32×32 grid patterns)
- Use colloidal probes or sharp tips depending on the specific property being measured
- Collect hundreds of force curves across the bacterial surface to capture heterogeneity [15]
Data Correlation: Correlate nanomechanical properties with biofilm formation assays (e.g., static microtiter plate assays) to establish structure-function relationships between capsular organization, adhesion forces, and biofilm formation capacity [15].
Visualization of Mechanical Mechanisms and Workflows
Diagram 1: Polysaccharide Mechanical Response Pathways. This diagram illustrates the relationship between molecular mechanisms, observable mechanical signatures, and characterization techniques used in polysaccharide nanomechanics studies.
Diagram 2: AFM Force Spectroscopy Experimental Workflow. This diagram outlines the standardized protocol for AFM-based nanomechanical characterization of polysaccharides, from sample preparation through data analysis.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 4: Essential Research Reagents for Polysaccharide Nanomechanics
| Reagent/Material | Function/Application | Specific Examples | Technical Considerations |
|---|---|---|---|
| AFM Cantilevers | Force detection and application | Soft cantilevers (0.01-0.1 N/m) | Requires precise spring constant calibration via thermal tuning [30] |
| Functionalized Substrates | Sample immobilization | Freshly cleaved mica, poly-L-lysine coated glass | Surface chemistry affects polysaccharide adsorption and conformation [29] [15] |
| Cholesterol-Conjugated Polysaccharides | Membrane functionalization | Chol-CS (Chondroitin Sulfate) | Enables asymmetric incorporation into outer leaflet of lipid bilayers [28] |
| Giant Unilamellar Vesicles (GUVs) | Model membrane systems | DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine) GUVs | Formed via electroformation; ideal for micropipette aspiration [28] |
| Fluorescent Labels | Visualization and tracking | FITC-labeled polysaccharides, lipid dyes | Enables confocal microscopy and FRAP measurements [28] |
| Microcapillary/Pipette Systems | Micropipette aspiration | Precision borosilicate glass capillaries | Requires precise pressure control systems for mechanical testing [28] |
The mechanical stability conferred by polysaccharides emerges from distinct biophysical mechanisms that can be quantitatively characterized using AFM nanomechanics and complementary biophysical techniques. Through entanglement networks, cross-linking junctions, helical bundle formations, and membrane interactions, polysaccharides generate identifiable mechanical signatures including sawtooth patterns, force plateaus, and force staircases. The experimental protocols and analytical frameworks presented herein provide researchers with standardized methodologies for investigating these phenomena, particularly in the context of biofilm formation and drug delivery system design. As research in this field advances, correlation of these nanomechanical properties with biological function will continue to reveal new opportunities for therapeutic intervention and biomaterial innovation.
Probing the Nanomechanical World: Advanced AFM Methodologies for Biofilm Analysis
Atomic Force Microscopy (AFM) is arguably the most versatile and powerful microscopy technique for nanoscale analysis, capable of achieving atomic resolution with Ångström-level height accuracy [31]. Unlike optical or electron microscopy, AFM does not use lenses or beam irradiation, and thus does not suffer from limitations in spatial resolution due to diffraction and aberration [32]. This technical guide explores the core principles of AFM, specifically topographical imaging and force spectroscopy, framed within its application to nanomechanics studies of capsular polysaccharides in biofilm research—a field crucial for understanding bacterial pathogenesis and developing novel therapeutic strategies.
Fundamental Operating Principles
Core Components and Sensing Mechanism
The fundamental operation of an AFM involves scanning a sample using a sharp tip (with a radius of curvature on the order of nanometers) mounted on a flexible cantilever [33] [32].
- Surface Sensing: As the tip approaches the sample surface, forces including attractive van der Waals and repulsive contact forces cause the cantilever to deflect. This delicate balance enables ultra-precise surface probing [31].
- Detection System: A laser beam reflects off the cantilever onto a position-sensitive photodetector (PSPD). Nanoscale deflections alter the laser's path, allowing the PSPD to track height variations and force interactions [31] [32].
- Imaging Mechanism: By scanning the tip across the sample while maintaining constant tip-sample interaction via a feedback loop, the AFM constructs a 3D topographic map with real-time quantitative data on properties such as adhesion and stiffness [31].
The following diagram illustrates the fundamental components and workflow of an AFM system:
Primary Imaging Modes
AFM operates primarily in three modes, each with distinct advantages for different sample types, including delicate biological specimens like bacterial capsules [31] [33].
Table 1: Key Characteristics of AFM Imaging Modes
| Operating Mode | Tip-Sample Interaction | Optimal For | Resolution | Risk of Sample Damage |
|---|---|---|---|---|
| Contact Mode | Constant physical contact [31] | Hard, rigid samples [31] | High (atomic resolution possible) [32] | High [33] |
| Tapping Mode | Intermittent contact (oscillating tip) [31] [33] | Soft, fragile, adhesive samples [31] | High (prevents lateral forces) [31] | Low [31] |
| Non-Contact Mode | No contact (attractive forces sensed) [31] | High-resolution in various environments [31] | Moderate [31] | Very Low [31] |
Force Spectroscopy
Principles and Methodology
Force spectroscopy transforms the AFM from a topographic imager into a sophisticated nanomechanical probe, directly measuring interaction forces between the tip and sample [32]. This is achieved by acquiring force-distance curves, which record the cantilever deflection as a function of the piezoelectric actuator's vertical position [31].
The experimental protocol involves:
- Approach Curve: The tip approaches the sample until contact is established, with repulsive forces causing cantilever bending.
- Retraction Curve: The tip withdraws from the surface, often revealing adhesive interactions through a characteristic "pull-off" event [31].
Analysis of these curves provides quantitative data on mechanical properties including adhesion force, elastic modulus (Young's modulus), and sample deformation [31] [32].
Application to Biofilm and Polysaccharide Research
In biofilm research, force spectroscopy is instrumental for measuring the mechanical properties of bacterial surfaces and their constituent polysaccharides. A key study on Klebsiella pneumoniae used AFM to perform in situ nanomechanical measurements of wild-type and mutant strains, revealing how capsular organization influences bacterial adhesion and biofilm formation [15]. Furthermore, a 2023 study correlated the intrinsic viscosity and electrokinetic properties of various capsular polysaccharides with their antibiofilm activity, measurements made possible by AFM-based force spectroscopy [16].
Table 2: Key Mechanical Properties Measurable via AFM Force Spectroscopy
| Property | Description | Derived From | Significance in Biofilm Research |
|---|---|---|---|
| Adhesion Force | Force required to separate tip from sample [31] | Retraction curve "pull-off" event [31] | Quantifies bacterium-surface and bacterium-bacterium interactions [15] |
| Young's Modulus | Measure of sample stiffness/elasticity [32] | Slope of the approach curve in the contact region [31] | Reveals mechanical robustness of bacterial capsules and biofilm matrix [15] [16] |
| Deformation | Degree of sample indentation under load | Difference between piezo movement and tip deflection | Informs on capsule compliance and its role in surface attachment [15] |
Advanced Techniques and Recent Developments
Correlative Microscopy and New Instrumentation
The integration of AFM with other analytical techniques is a powerful trend. Correlative systems combine nanometre topographical information from AFM with optical and spectral chemical information from techniques like fluorescence microscopy, enabling the linking of properties at the nanoscale [34]. New instruments, such as Oxford Instruments Asylum Research's Cypher Vero with Quadrature Phase Differential Interferometry (QPDI) for more accurate tip displacement sensing, are pushing technological barriers, particularly for techniques like Piezoresponse Force Microscopy (PFM) [34].
AI and Data Analysis
The application of Artificial Intelligence (AI) and Machine Learning (ML) is growing within AFM operation and data analysis. These technologies are being used to train algorithms for probe inspection, image processing, and to analyze complex, multi-parametric data (e.g., mechanical, optical) to identify trends and relationships that are difficult to discern manually [34]. A recent development is AFMfit, an open-source Python package that uses a fast fitting algorithm based on nonlinear Normal Mode Analysis to interpret conformational dynamics in AFM data, processing hundreds of images in minutes [35].
The Scientist's Toolkit: AFM in Biofilm Research
Table 3: Essential Research Reagent Solutions for AFM Nanomechanics of Biofilms
| Item / Reagent | Function / Role | Application Example |
|---|---|---|
| Conductive AFM Probes | Measure current flow for electrical characterization (C-AFM) [31] | Mapping local conductivity of extracellular polymeric substances |
| Sharp Silicon Nitride Tips | High-resolution topographical imaging of soft biological samples [33] [32] | Visualizing polysaccharide fiber networks in biofilms |
| Functionalized Tips | Tips coated with specific molecules (e.g., lectins, antibodies) [32] | Probing specific receptor-ligand interactions on bacterial surfaces |
| Liquid Imaging Cell | Enables AFM operation in physiological buffers [31] | In-situ nanomechanics of living bacteria in biofilm conditions |
| Purified Capsular Polysaccharides | Reference samples for structure-function studies [16] | Identifying biophysical properties (e.g., intrinsic viscosity) that confer antibiofilm activity [16] |
The following diagram summarizes the integrated experimental workflow for an AFM nanomechanics study of biofilms, from sample preparation to data interpretation:
In Situ Nanomechanical Profiling of Live Biofilms
Atomic force microscopy (AFM) has emerged as a powerful tool for interrogating the structural and mechanical properties of live biofilms under physiological conditions. This technical guide details the application of AFM nanomechanics to profile the dynamic architecture of biofilms, with a specific focus on the role of capsular polysaccharides. We provide comprehensive protocols for in-situ imaging and force measurements, quantitative data on bacterial mechanical properties, and advanced methodologies for large-area analysis. Within the broader thesis of AFM nanomechanics of capsular polysaccharides, this work illuminates how biophysical properties govern biofilm assembly and resistance, offering new avenues for anti-biofilm strategies in therapeutic development.
Biofilms are structured microbial communities encased in a self-produced extracellular polymeric substance (EPS) that confers significant tolerance to antimicrobial agents and host defenses [36] [5]. The biofilm matrix is a complex amalgam of polysaccharides, proteins, nucleic acids, and other biomolecules that determine the mechanical integrity and resilience of the biofilm [5]. Atomic force microscopy (AFM) has transitioned from a surface imaging technique to a sophisticated nanomechanical probe capable of quantifying the biophysical properties of live biofilms in their native, hydrated state [37]. This capability is critical for understanding biofilm pathogenesis and developing effective countermeasures.
The application of AFM nanomechanics to the study of capsular polysaccharides provides a unique window into the fundamental mechanisms of biofilm assembly and stability. These polysaccharides are key components of the bacterial capsule and EPS, influencing adhesion, cohesion, and overall biofilm architecture [15] [16]. By measuring properties such as Young's modulus, turgor pressure, and adhesion forces at the single-cell and community levels, researchers can decipher the structure-function relationships of polysaccharides without relying on biocidal agents or disruptive sample preparation [38] [16]. This guide details the experimental frameworks and analytical methods for obtaining such crucial nanomechanical profiles.
Key Nanomechanical Properties and Measurements
AFM force-distance curves are the fundamental source for quantifying the nanomechanical signature of biofilms. The figure below outlines the core workflow for data acquisition and analysis.
Figure 1: AFM Force-Distance Curve Analysis Workflow. The process begins with the tip approaching and retracting from the cell surface, generating approach and retraction curves. These curves are analyzed to extract key nanomechanical properties.
The following table summarizes critical nanomechanical properties that can be derived from AFM force spectroscopy and their biological significance in the context of biofilms and capsular polysaccharides.
Table 1: Key Nanomechanical Properties Measured by AFM in Biofilm Studies
| Property | Description | Typical AFM Measurement | Biological Significance in Biofilms |
|---|---|---|---|
| Young's Modulus | Measures stiffness or elastic resistance to deformation [37]. | Derived from slope of the linear compression regime in approach curve, often using Hertz model [38] [37]. | Indicates cell wall rigidity; influenced by polysaccharide capsule and EPS composition [15]. |
| Turgor Pressure | Internal hydrostatic pressure within the cell [38]. | Calculated from force-indentation data and cell wall deformation models. | Critical for cell integrity; varies with gliding motility and environmental conditions [38]. |
| Adhesion Force | Strength of attractive forces between the AFM tip (or modified probe) and the sample [39] [37]. | Measured as the maximum pull-off force in the retraction curve [39] [37]. | Quantifies stickiness of bacterial surface or EPS; key for initial surface attachment and biofilm cohesion [39]. |
| Cell Stiffness (k~cell~) | Resistance of the entire cell body to compression [37]. | Calculated from the effective spring constant (k~effective~) in the linear regime of the approach curve [37]. | Reflects overall mechanical stability of the cell; can be altered in biofilm-specific phenotypes. |
Quantitative Profiling of Bacterial Mechanics
AFM studies have successfully quantified the mechanical properties of diverse bacterial species. For instance, in-situ profiling of live, gliding Nostoc cyanobacteria and non-motile Rhodococcus wratislaviensis under physiological conditions revealed Young's modulus values ranging from 20 ± 3 MPa to 105 ± 5 MPa, and turgor pressures from 40 ± 5 kPa to 310 ± 30 kPa, depending on the bacterium and its gliding speed [38]. These measurements were made possible by an AFM procedure based on fast force-distance curves at every pixel, which eliminates the need for chemical or mechanical immobilization and reduces lateral forces [38].
Furthermore, nanomechanical measurements of the pathogen Klebsiella pneumoniae have demonstrated that the structural organization of the capsular polysaccharide capsule directly influences bacterial adhesion and the initial stages of biofilm formation [15]. The adhesion forces between a single bacterial cell and a surface, quantified using AFM, typically fall in the nanonewton range. For example, studies on sulfate-reducing bacteria (SRB) reported adhesion forces between the AFM tip and the cell surface in the range of -3.9 nN to -4.3 nN [39].
Experimental Protocols for In-Situ Profiling
Sample Preparation with Minimal Perturbation
A critical step for in-situ nanomechanical profiling is the immobilization of live biofilms or bacterial cells without compromising their viability or native state.
- Non-Immobilization Method for Motile Bacteria: For gliding or motile bacteria like Nostoc, a gentle sample preparation process combined with an AFM procedure using fast force-distance curves at every pixel can be employed. This method drastically reduces lateral forces, allowing imaging and mechanical profiling without any external immobilization protocol [38].
- Physical Entrapment: Non-motile cells can be mechanically trapped in porous membranes like isopore polycarbonate or polydimethylsiloxane (PDMS) stamps [37]. This method avoids chemical fixation and is suitable for spherical cells, though it may impose some mechanical stress.
- Biofilm Growth on Substrata: Growing cells as biofilms directly on glass coverslips or other suitable surfaces leverages their natural adhesive properties, eliminating the need for external fixatives. This approach is highly physiologically relevant, though the EPS layer will be included in the measurements [37].
- Electrostatic Adhesion: For planktonic cells, the most common method involves treating glass coverslips with polycationic agents like poly-L-lysine or Corning Cell-Tak to create a positively charged surface that strongly adheres the negatively charged bacterial cells [37]. Cell-Tak often provides more robust adhesion.
AFM Force Spectroscopy Protocol
The following protocol details the steps for obtaining nanomechanical profiles from live biofilms.
- Cantilever Calibration: Before force measurement, calibrate the cantilever's spring constant (k~cantilever~) on a clean, hard surface (e.g., bare glass or silicon wafer) in fluid [37]. This is a prerequisite for quantifying all absolute force values.
- System Setup: Perform all measurements in appropriate fluid cells using the bacterial growth medium or a compatible buffer to maintain physiological conditions and avoid capillary forces from air-liquid interfaces [37].
- Tip Selection: Use sharp, non-functionalized tips for topographical imaging and basic mechanical property mapping. For specific adhesion studies (e.g., polysaccharide-ligand interactions), the tip can be functionalized with relevant molecules [37].
- Force Volume Imaging: Acquire force-distance curves at every pixel of a defined grid over the biofilm surface. This mode simultaneously generates a topographical image and a map of mechanical properties [38].
- Data Acquisition Parameters:
- Set Point: Choose a maximum loading force that is sufficient to induce a measurable indentation but low enough to avoid damaging the cell (typically 0.5-10 nN) [37].
- Approach/Retraction Speed: Optimize speed to capture the relevant polymer dynamics; slower speeds (e.g., 0.5-1 µm/s) can provide more detail on polymer unfolding and viscoelastic responses.
- Spatial Resolution: The density of force curves should be chosen based on the spatial heterogeneity of the biofilm. High-resolution maps may require a grid of 128x128 or 256x256 points.
Data Analysis and Model Fitting
- Elasticity and Stiffness: Fit the approach curve's nonlinear compression region with the Hertz model to calculate Young's modulus [37]. Calculate the cell stiffness (k~cell~) from the slope of the linear compression regime and the known k~cantilever~ [37].
- Adhesion: Analyze the retraction curve to identify the maximum pull-off force (adhesion force) and the work of adhesion (area under the retraction curve) [39] [37]. The presence of multiple rupture events often indicates the unbinding of multiple polymer chains or adhesins.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials and Reagents for AFM Biofilm Nanomechanics
| Item | Function/Application | Examples & Notes |
|---|---|---|
| AFM with Fluid Cell | Enables imaging and force measurement under physiological liquid conditions [38] [37]. | Must support force-volume or peak-force quantitative nanomechanical (PF-QNM) modes. |
| Poly-L-Lysine | Coating agent to create a positively charged surface for electrostatic immobilization of bacterial cells [37]. | A common, but not always the most robust, adhesion method. |
| Corning Cell-Tak | Bio-adhesive for robust immobilization of cells to the AFM substrate [37]. | Can provide more reliable adhesion than poly-L-lysine for certain organisms. |
| Polycarbonate Membranes / PDMS Stamps | For physical entrapment of cells, avoiding chemical treatments [37]. | Ideal for yeast or non-motile bacteria where chemical gluing is undesirable. |
| Functionalization Kits | For coating AFM tips with specific ligands, antibodies, or molecules to study specific interactions [37]. | Allows probing of specific polysaccharide-protein interactions. |
| Hertz Contact Model | Analytical model used to derive Young's modulus from force-indentation data [37]. | Assumes an isotropic, linear elastic material; use with caution for highly heterogeneous EPS. |
Advanced Applications: Capsular Polysaccharides and Large-Area Analysis
Nanomechanics of Capsular Polysaccharides
Capsular polysaccharides are not merely passive physical barriers; they are active mediators of biofilm mechanics with distinct electrokinetic signatures. A key finding is that antibiofilm capsular polysaccharides, which non-biocidally prevent adhesion and biofilm formation, share common biophysical properties. Specifically, all active macromolecules display high intrinsic viscosity [η] (> 7 dl/g) and a distinct electrokinetic signature characterized by a high density of electrostatic charges [16]. This high intrinsic viscosity reflects a expanded, hydrated conformation in solution that is critical for their anti-adhesion activity, as even minor size reduction of these polymers leads to a loss of function [16]. AFM nanomechanics can probe how these polymers alter the cell surface properties and interface with surfaces.
Large-Area AFM and Machine Learning Integration
A major limitation of conventional AFM in biofilm research is the small imaging area (<100 µm), which makes it difficult to link nanoscale properties to the millimeter-scale architecture of biofilms [5]. This limitation is now being addressed by automated large-area AFM, which can capture high-resolution images over millimeter-scale areas [5]. This approach reveals spatial heterogeneity and cellular morphology during early biofilm formation that was previously obscured.
The workflow involves automated acquisition of multiple contiguous AFM images, which are then seamlessly stitched together. To manage the resulting large, information-rich datasets, machine learning (ML) algorithms are implemented for image segmentation, cell detection, and classification [5]. This integration allows for the quantitative analysis of parameters such as cell count, confluency, shape, and orientation over vast areas, providing unprecedented insights into biofilm organization and the role of structures like flagella in community assembly [5].
Figure 2: Workflow for Large-Area AFM Analysis of Biofilms. The process involves automated tiled scanning, image stitching, and machine learning-based analysis to generate quantitative maps of biofilm architecture across millimeter scales.
In-situ nanomechanical profiling with AFM provides an unparalleled, quantitative description of the biophysical landscape of live biofilms. The methodologies outlined herein—from fundamental force spectroscopy to advanced large-area mapping—enable researchers to decipher the mechanical contributions of key matrix components like capsular polysaccharides. The integration of machine learning and automation is poised to further transform the field, allowing for the correlation of nanoscale properties with macroscale biofilm behavior. For researchers and drug development professionals, these tools and insights are critical for designing novel, mechanism-based strategies to combat biofilm-associated infections, moving beyond traditional biocidal approaches to target the very physical foundations of biofilm resilience.
Atomic Force Microscopy (AFM) has emerged as a powerful tool in biofilm research, enabling nanoscale topographical and nanomechanical characterization under physiological conditions without extensive sample preparation [40]. This case study employs AFM nanomechanics to investigate the role of capsular polysaccharides (CPS) in biofilm formation and mechanical properties of Klebsiella pneumoniae, a significant human pathogen. The capsule, a thick polysaccharide layer surrounding the cell, is a major virulence factor, but its nanomechanical role in biofilm development remains incompletely understood [24]. By comparing wild-type and isogenic capsular mutants, this research provides quantitative insights into how CPS influences bacterial adhesion, surface organization, and resistance to environmental stresses, offering potential pathways for targeted anti-biofilm strategies.
Background
Klebsiella pneumoniae and Biofilm-Associated Infections
Klebsiella pneumoniae is a gram-negative pathogen responsible for healthcare-associated infections including pneumonia, urinary tract infections, and bacteremia. Its pathogenicity is heavily linked to the formation of biofilms—structured communities of bacterial cells enclosed in an extracellular polymeric matrix—on both biotic surfaces and abiotic medical devices [24]. Biofilms confer enhanced resistance to antibiotics and host immune responses, making infections difficult to eradicate.
AFM in Biofilm Research
Traditional imaging techniques like scanning electron microscopy (SEM) require extensive sample preparation, including dehydration and metal coating, which can alter native biofilm structures [40]. AFM overcomes these limitations by operating in ambient conditions or liquid environments, preserving the sample's native state [5]. AFM provides unprecedented resolution for visualizing:
- Individual bacterial cells and their surface appendages (e.g., flagella, pili)
- Extracellular Polymeric Substances (EPS) that form the biofilm matrix
- Nanomechanical properties including stiffness, adhesion, and viscoelasticity [5] [24]
Recent advancements, such as automated large-area AFM combined with machine learning for image stitching and analysis, now enable researchers to capture high-resolution data over millimeter-scale areas, linking cellular-scale features to the functional architecture of entire biofilm communities [5].
Methodology
Bacterial Strains and Growth Conditions
Wild-type (WT) Klebsiella pneumoniae and specific isogenic capsular mutants were cultivated in appropriate liquid growth media to mid-logarithmic phase. Mutants were generated through targeted gene disruption of key polysaccharide synthesis genes (e.g., wza, wzc) to produce acapsular or reduced-capsule variants [24].
Sample Preparation for AFM
- Surface Attachment: Bacterial suspensions were deposited onto freshly cleaved mica or PFOTS-treated glass substrates [5].
- Gentle Rinsing: Unattached cells were removed by carefully rinsing with ultrapure water or phosphate-buffered saline (PBS) to preserve weakly adherent structures.
- Imaging Conditions: Experiments were performed in both ambient conditions and liquid (PBS) to assess topological and mechanical properties under near-physiological conditions [24] [40].
Atomic Force Microscopy Operation
A comprehensive overview of key research reagents and materials is provided in the table below:
Table 1: Research Reagent Solutions and Essential Materials
| Item Name | Function/Application |
|---|---|
| Klebsiella pneumoniae Wild-Type | Reference strain for comparing structural and mechanical properties with mutants [24]. |
| Capsular Mutants (e.g., ΔcapD) | Isogenic mutants with deficient capsule synthesis; crucial for elucidating the specific role of CPS [24]. |
| PFOTS-Treated Glass | Hydrophobic surface treatment used to promote and study bacterial adhesion under controlled conditions [5]. |
| Mica Substrates | Atomically flat surfaces ideal for high-resolution AFM imaging of bacterial adhesion and topography [24]. |
| Sharp Nitride Lever AFM Probes | Probes with high resonance frequencies for topographical imaging and nanomechanical property mapping [24]. |
AFM Imaging Modes:
- Contact Mode: Used for high-resolution topographical imaging of surface structures.
- Force Spectroscopy: Employed to quantitatively map nanomechanical properties including stiffness (Young's modulus) and adhesion forces by collecting force-distance curves at multiple locations across the bacterial surface [24].
Large-Area AFM Protocol:
- Automated Scanning: Pre-defined millimeter-sized areas were scanned using an automated stage to collect multiple contiguous high-resolution images [5].
- Image Stitching: Machine learning algorithms were employed to seamlessly stitch individual images into a cohesive large-area map with minimal overlap between scans [5].
- Data Analysis: Machine learning-based segmentation automatically extracted quantitative parameters including cell count, confluency, cell shape, and orientation from the large-area AFM data [5].
Data Analysis and Theoretical Modeling
- Young's Modulus Calculation: Force-distance curves were analyzed using Hertzian contact mechanics models to calculate Young's modulus, a measure of cell stiffness [24].
- Adhesion Force Measurement: The maximum pull-off force in retraction curves was quantified to assess adhesion between the AFM tip and the bacterial surface.
- Theoretical Modeling: The organization of the capsular polysaccharide layer and its influence on bacterial adhesion was interpreted through theoretical polymer hydrogel models [24].
Results and Discussion
Nanomechanical Properties of Capsular Polysaccharides
AFM force spectroscopy revealed the CPS of wild-type K. pneumoniae behaves as a responsive polymer hydrogel. The capsule undergoes significant compression under applied force, demonstrating its role as a mechanical buffer [24] [23].
Key quantitative findings from nanomechanical measurements are summarized below:
Table 2: Quantitative Nanomechanical Properties of K. pneumoniae Strains
| Strain | Young's Modulus (Stiffness) | Adhesion Force | Capsule Height | Key Structural Feature |
|---|---|---|---|---|
| Wild-Type | Low (Soft) | Moderate | ~400 nm | Organized, dense polysaccharide matrix |
| Capsular Mutant | High (Stiff) | High / Low (variable) | Not detectable | Absence of capsule layer |
This data indicates the wild-type capsule's soft, compliant nature directly reduces adhesion forces by preventing close contact between the AFM tip (or surface) and the rigid cell wall. In contrast, capsular mutants, lacking this hydrogel layer, exhibit higher stiffness and altered adhesion profiles [24]. This provides a nanomechanical basis for the capsule's role in mitigating interactions with surfaces and potentially with antimicrobial agents.
Role of CPS in Biofilm Architecture and Adhesion
Large-area AFM imaging demonstrated that wild-type K. pneumoniae forms robust biofilms with a confluent cellular network. The CPS facilitates a repulsive, soft interface between adjacent cells, promoting the formation of a porous, three-dimensional structure [24].
Capsular mutants, however, showed aberrant biofilm morphology. Without the protective capsule, mutants often formed denser, flatter biofilms with increased cell-to-cell and cell-to-substrate contact, corroborating the higher adhesion forces measured by force spectroscopy [24]. This suggests the capsule is critical for establishing the optimal spatial organization for mature biofilm development, not just for initial attachment.
CPS as an Adaptive Hydrogel for Osmotic Protection
Beyond adhesion modulation, AFM studies demonstrated the capsule's function as an adaptive hydrogel. In hyperosmotic conditions, the wild-type capsule dehydrates and collapses, reducing its volume and height while increasing its stiffness. This reversible process helps maintain cell turgor pressure and viability [23]. The polysaccharide matrix acts as an "ion sponge," dampening the impact of osmotic stress on the cell proper. This mechanism was absent in capsular mutants, which showed no such adaptive response and suffered greater physiological damage under osmotic shock [23].
Diagram 1: CPS osmotic stress response
This AFM nanomechanics case study establishes that the capsular polysaccharide of Klebsiella pneumoniae is not merely a static protective barrier but a dynamic, responsive polymer hydrogel crucial for biofilm pathophysiology. The capsule directly modulates key mechanical properties—reducing cellular stiffness and adhesion—to facilitate the formation of architecturally complex biofilms. Furthermore, its ability to adaptively collapse under osmotic stress provides a novel mechanical mechanism for cell protection. These findings underscore the importance of targeting CPS assembly and mechanical properties in the development of novel anti-biofilm strategies against this resilient pathogen.
Diagram 2: AFM experimental workflow
Automated Large-Area AFM and Machine Learning for Millimeter-Scale Analysis
Atomic force microscopy (AFM) is a powerful tool in biofilm research, providing nanoscale resolution of topographical features and nanomechanical properties of capsular polysaccharides under physiological conditions [41] [42]. These exopolysaccharides form a critical component of the extracellular polymeric substance (EPS) matrix, mediating surface attachment, mechanical stability, and antimicrobial resistance in biofilms [20]. Traditional AFM faces significant limitations in studying biofilm systems due to its restricted scan range (typically <100 µm), labor-intensive operation, and inability to capture the millimeter-scale spatial heterogeneity inherent to microbial communities [42].
The integration of automated large-area AFM with machine learning (ML) frameworks now enables comprehensive analysis of biofilm assembly across multiple scales [42] [43]. This technical guide outlines methodologies and applications of these advanced techniques specifically for investigating the role of capsular polysaccharides in biofilm formation and nanomechanics, providing researchers with protocols to bridge the gap between nanoscale biophysical properties and macroscale community organization.
Technical Foundations of Automated Large-Area AFM
System Components and Configuration
Automated large-area AFM systems overcome traditional range limitations through integrated hardware and software solutions. The core components include:
- Extended Range Nanopositioning Stages: Systems utilizing position-velocity-time control with pre-programmed coordinates enable high-speed (up to 3 mm/s) scanning over areas up to 100 µm × 100 µm, with coarse positioning extending effective range to 0.5 mm × 0.7 mm through image stitching [44].
- Python-based Automation Interfaces: Application Programming Interfaces (APIs) allow full scripted control of AFM operations, enabling automated multi-region imaging and continuous, unsupervised data acquisition over extended durations [43].
- Machine Learning Integration: ML frameworks implement autonomous sample selection, probe conditioning assessment, and real-time image quality evaluation to reduce human supervision and improve data consistency [41] [42].
ML-Enhanced Data Acquisition Workflow
The integration of machine learning transforms the AFM workflow from manual operation to an automated, intelligent system for large-area biofilm characterization, as illustrated below:
Experimental Protocols for Capsular Polysaccharide Characterization
Large-Area AFM Imaging of Biofilm Assembly
Objective: To characterize the spatial organization and structural role of capsular polysaccharides during early biofilm formation across millimeter-scale areas.
Sample Preparation Protocol:
- Surface Functionalization: Treat glass coverslips with PFOTS (1H,1H,2H,2H-Perfluorooctyltriethoxysilane) to create a hydrophobic surface that promotes bacterial attachment while minimizing non-specific binding [42].
- Bacterial Immobilization: Inoculate Pantoea sp. YR343 (or target strain) in liquid growth medium and transfer to Petri dishes containing functionalized coverslips.
- Controlled Incubation: Incubate for specific durations (e.g., 30 minutes for initial attachment studies; 6-8 hours for cluster formation) under appropriate physiological conditions.
- Sample Fixation: Gently rinse coverslips to remove unattached cells and air-dry before AFM imaging to preserve native structures [42].
AFM Imaging Parameters:
- Mode: Tapping Mode or PeakForce Tapping for minimal sample disturbance [45]
- Scan Rate: 0.5-1.0 Hz for high-resolution images
- Resolution: 512 × 512 pixels per individual scan
- Scan Size: 10 µm × 10 µm to 100 µm × 100 µm per tile
- Cantilever Selection: Sharp tips (nominal radius <10 nm) with spring constant of ~0.4 N/m for optimal resolution of polysaccharide structures [42]
Large-Area Automation:
- Program coordinate grid for automated tiling across millimeter-scale areas using Python scripting interface [43].
- Implement minimal overlap (5-10%) between adjacent tiles to maximize acquisition speed while enabling seamless stitching.
- Utilize ML-based image assessment to flag and rescan regions with quality issues (e.g., tip artifacts, drift) during acquisition [41].
Nanomechanical Property Mapping of Capsular Polysaccharides
Objective: To quantitatively measure the mechanical properties of capsular polysaccharides and their relationship to biofilm architecture.
PeakForce QNM Protocol:
- Calibration: Pre-calibrate cantilever spring constant using thermal tune method and determine optical lever sensitivity on rigid reference sample (e.g., silicon wafer) [45].
- Parameter Optimization: Set PeakForce frequency to 0.5-2 kHz and amplitude to 50-100 nm to ensure sufficient force sensitivity while avoiding sample damage.
- Data Collection: Acquire force-distance curves at each pixel (typically 256 × 256 or 512 × 512 resolution) with maximum applied force of 1-5 nN for biological samples.
- Model Fitting: Apply appropriate contact mechanics models (DMT, Hertz) to derive quantitative mechanical properties [45].
ML-Enhanced Data Analysis:
- Implement clustering algorithms (K-Means, Gaussian Mixture Models) to automatically identify and segment regions with distinct mechanical properties corresponding to different EPS components [45].
- Apply convolutional neural networks (CNNs) trained on known polysaccharide features to classify structures and map their distribution across large areas [41].
Table 1: Key Nanomechanical Properties of Biofilm Components
| Biofilm Component | Reduced Modulus (MPa) | Adhesion (nN) | Deformation (nm) | Measurement Technique |
|---|---|---|---|---|
| Capsular Polysaccharides | 0.5-5.0 | 0.1-0.5 | 1-5 | PeakForce QNM [15] [45] |
| Bacterial Cell Wall | 10-100 | 0.5-2.0 | 0.5-2 | AFM Nanomechanical Mapping [42] |
| Flagellar Structures | 1-3 | 0.2-0.8 | 2-8 | High-Resolution Force Spectroscopy [42] |
| Mature EPS Matrix | 0.1-1.0 | 0.5-3.0 | 5-20 | AFM-nDMA [45] |
Machine Learning Applications for Data Analysis
Image Stitching and Segmentation
The massive datasets generated by large-area AFM require automated processing pipelines to extract biologically meaningful information:
- Seamless Stitching Algorithm: ML-based feature matching enables accurate tile alignment even with minimal (5-10%) overlap between scans, compensating for stage drift and image distortions [42].
- CNN-Based Segmentation: U-Net architectures trained on manually annotated AFM images automatically identify and segment individual bacterial cells, flagellar structures, and EPS regions with >90% accuracy [42].
- Morphological Analysis: Automated quantification of cell dimensions, orientation, surface coverage (confluency), and spatial distribution patterns across millimeter scales [42].
Structure-Function Relationship Analysis
Machine learning enables correlation of structural features with nanomechanical properties to understand polysaccharide function:
- Regression Models: Predict mechanical properties (adhesion, stiffness) from topological features using random forest or gradient boosting algorithms.
- Dimensionality Reduction: t-SNE or UMAP visualization of high-dimensional nanomechanical data to identify distinct compositional regions within heterogeneous biofilms [45].
- Time-Series Analysis: Recurrent neural networks (RNNs) track biofilm development dynamics from sequential large-area scans acquired over hours to days [42].
Table 2: Quantitative Parameters from Large-Area AFM of Pantoea sp. YR343 Biofilms
| Morphological Parameter | 30-Minute Attachment | 6-8 Hour Cluster Formation | Measurement Method |
|---|---|---|---|
| Cell Length (µm) | 1.8-2.2 | 1.9-2.3 | Automated ML Segmentation [42] |
| Cell Diameter (µm) | 0.9-1.1 | 0.9-1.2 | Automated ML Segmentation [42] |
| Flagellar Height (nm) | 20-50 | 20-50 | High-Resolution Topography [42] |
| Spatial Organization | Isolated cells | Honeycomb pattern | Spatial Autocorrelation Analysis [42] |
| Surface Coverage (%) | 5-15 | 25-40 | Pixel-based ML Classification [42] |
Research Reagent Solutions and Essential Materials
Table 3: Essential Research Reagents and Materials for AFM Biofilm Studies
| Item | Function/Application | Specifications |
|---|---|---|
| PFOTS-Treated Substrates | Hydrophobic surface for controlled bacterial attachment | Glass coverslips functionalized with 1H,1H,2H,2H-Perfluorooctyltriethoxysilane [42] |
| Nanopositioning Stage | Extended range scanning for large-area acquisition | Queensgate NPS-XY-100D or equivalent with 100 µm × 100 µm range [44] |
| Sharp AFM Probes | High-resolution imaging of polysaccharide structures | Cantilevers with nominal tip radius <10 nm, spring constant ~0.4 N/m [42] |
| Python AFM Library | Automation and scripted control of AFM operations | Nanosurf Python API or equivalent for automated multi-region imaging [43] |
| ML Segmentation Tools | Automated cell detection and morphological analysis | U-Net or similar CNN architectures trained on AFM image datasets [42] |
Case Study: Nanomechanics of Klebsiella pneumoniae Capsular Polysaccharides
The relationship between capsular polysaccharides and biofilm formation can be investigated through targeted nanomechanical analysis:
Experimental Design:
- Compare wild-type K. pneumoniae with isogenic mutants deficient in capsule synthesis or fimbriae production [15].
- Perform large-area AFM imaging to characterize spatial distribution and organization of capsular polysaccharides.
- Apply PeakForce QNM to measure adhesion forces and reduced modulus of individual cells and surrounding EPS matrix.
Key Findings:
- Wild-type strains with organized capsular architecture show enhanced adhesion and biofilm formation capacity [15].
- Mutants with disrupted capsule organization exhibit significantly reduced adhesion forces (30-50% decrease) despite similar chemical composition [15].
- Type 3 fimbriae directly influence capsular organization, demonstrating the interplay between different surface structures in biofilm development [15].
The relationship between polysaccharide biophysical properties and antibiofilm activity can be systematically characterized as shown below:
Table 4: Biophysical Properties of Antibiofilm Capsular Polysaccharides
| Polysaccharide | Molecular Weight (kDa) | Intrinsic Viscosity (dl/g) | Antibiofilm Spectrum | Key Structural Features |
|---|---|---|---|---|
| G2cps (E. coli) | 800 | >15 | Broad (Gram+/Gram-) | Glycerol phosphate repeats, O-acetylated [16] |
| Vi (S. Typhi) | 350 | 12.5 | Broad (Gram+/Gram-) | α-1,4-linked GalNAcA, O-acetylated [16] |
| MenA | 220 | 9.8 | Broad (Gram+/Gram-) | →6)-α-D-ManpNAc(3OAc)-(1→PO4→ [16] |
| PnPS3 | 300 | 8.2 | Broad (Gram+/Gram-) | [→3)-β-D-Glcp-(1→3)-α-D-GlcpA-(1→]n [16] |
| PnPS18C | 180 | 6.5 | Narrow (S. aureus only) | [→4)-β-D-Glcp-(1→4)-β-D-Glcp-(1→4)-β-D-Glcp-(1→]n [16] |
The integration of automated large-area AFM with machine learning frameworks represents a transformative approach for investigating capsular polysaccharides in biofilm systems. These methodologies enable researchers to bridge critical scale gaps, correlating nanomechanical properties of individual polysaccharides with their organizational role in millimeter-scale biofilm architecture. The protocols and analyses outlined in this technical guide provide a foundation for advanced studies of biofilm mechanics, with significant implications for developing anti-biofilm strategies targeting capsular polysaccharide function and assembly.
Correlating Nanomechanical Data with Phenotypic Biofilm Assays
The study of bacterial biofilms is critically important in biomedical research, particularly in the context of chronic infections and antimicrobial resistance (AMR). A key structural and functional component of biofilms is the extracellular polymeric substance (EPS), a matrix in which capsular polysaccharides are embedded, providing mechanical stability and protection to the resident microbial cells [20]. Traditional phenotypic biofilm assays, such as the crystal violet microtiter plate-based method, provide valuable information on biofilm formation capacity but offer limited insight into the nanoscale mechanical properties that govern biofilm function and resilience [46]. Atomic force microscopy (AFM) has emerged as a powerful tool to address this gap, enabling the quantification of mechanical properties of biofilm matrices with picoNewton (pN) scale sensitivity under physiologically relevant conditions [47] [48].
The integration of AFM nanomechanical data with conventional phenotypic assays represents a transformative approach in biofilm research. This correlative methodology allows researchers to connect macroscopic biofilm phenotypes with the nanomechanical properties of their constituent polysaccharides, offering unprecedented insights into structure-function relationships. Such correlations are particularly valuable for understanding the role of specific exopolysaccharides—including PNAG (Poly-β-(1→6)-N-acetylglucosamine), Alginate, Psl, and Pel—in biofilm development, stability, and resistance mechanisms [20]. This technical guide provides researchers with the methodological framework and experimental protocols necessary to successfully implement this integrated approach, with particular emphasis on its application within the broader context of AFM nanomechanics studies of capsular polysaccharides.
Theoretical Foundations: Biofilm Polysaccharides and Nanomechanics
Key Exopolysaccharides in Biofilm Formation
The biofilm matrix is composed of various exopolysaccharides that serve distinct structural and functional roles throughout the biofilm lifecycle. Understanding the biochemical properties of these polymers is essential for interpreting nanomechanical data:
PNAG (Polysaccharide Intercellular Adhesin): This linear homopolysaccharide consists of β(1,6)-linked N-acetylglucosamine residues, with approximately 10-20% of the amino groups non-acetylated in its mature form [20]. The partial deacetylation imparts a positive charge, enabling association with negatively charged bacterial cell surfaces—a critical property for biofilm formation and immune evasion. In staphylococcal species, PNAG is synthesized by proteins encoded by the icaADBC operon and has been demonstrated to be essential for biofilm formation under high-shear flow conditions [20].
Alginate, Psl, and Pel: Pseudomonas aeruginosa, a model organism for biofilm studies, produces three distinct exopolysaccharides: Alginate, Psl, and Pel [20]. Alginate is a polyanionic copolymer of β-D-mannuronate and its C-5 epimer α-L-guluronate, while Psl is a pentasaccharide repeating unit containing D-mannose, D-glucose, and L-rhamnose. Pel is a cationic glucose-rich polysaccharide that requires c-di-GMP for its production. Each polysaccharide is associated with different stages of P. aeruginosa biofilm development, with Psl and Pel involved in initial attachment and Alginate contributing to the structural integrity of mature biofilms [20].
Nanomechanical Properties of Biofilm Polysaccharides
AFM enables the quantification of several key nanomechanical properties that characterize biofilm polysaccharides:
Young's Modulus (Elasticity): This property describes the stiffness of the biofilm matrix and its constituent polysaccharides. Typical values for bacterial cells range from 200-300 kPa at cell edges to 1-1.5 MPa at the center, though these values can vary significantly depending on environmental conditions, growth phase, and polysaccharide composition [49]. Changes in Young's modulus often reflect structural rearrangements within the biofilm matrix in response to environmental stressors.
Adhesion Forces: AFM force spectroscopy measures the tip-sample interaction forces, providing insights into the adhesive properties of the biofilm surface. These measurements can reveal molecular interactions between AFM tips (functionalized with specific ligands) and polysaccharide components, with typical adhesion forces for bacterial surfaces ranging from nanonewtons to micronewtons depending on the imaging media and surface molecules [47] [49].
Surface Roughness: Topographical imaging at nanoscale resolution enables the quantification of surface roughness parameters, which correlate with biofilm heterogeneity and can indicate sites of active EPS secretion or structural rearrangement in response to environmental cues [49].
Table 1: Key Exopolysaccharides in Biofilm Formation and Their Properties
| Polysaccharide | Monomer Composition | Charge Characteristics | Primary Functions in Biofilm | Representative Producing Organisms |
|---|---|---|---|---|
| PNAG/PIA | β(1,6)-linked N-acetylglucosamine (partially deacetylated) | Cationic (~15% non-acetylated) | Cell-surface adhesion, inter-cellular aggregation, immune evasion | Staphylococcus aureus, Staphylococcus epidermidis, Escherichia coli |
| Alginate | β-D-mannuronate and α-L-guluronate | Polyanionic | Biofilm architecture, stability, protection against antibiotics | Pseudomonas aeruginosa |
| Psl | D-mannose, D-glucose, L-rhamnose | Neutral to anionic | Surface attachment, initial microcolony formation, biofilm scaffolding | Pseudomonas aeruginosa |
| Pel | N-acetylgalactosamine, N-acetylglucosamine | Cationic | Cell-cell adhesion, structural integrity, cation sequestration | Pseudomonas aeruginosa |
Methodological Framework: Correlative AFM and Phenotypic Assays
Integrated Experimental Workflow
The successful correlation of nanomechanical data with phenotypic biofilm assays requires a systematic, multi-stage approach. The following workflow diagram illustrates the integrated experimental pipeline:
Experimental Protocols
Sample Preparation for Correlative Microscopy
Biofilm Growth and Immobilization:
- Cultivate biofilms on appropriate substrates (e.g., glass coverslips, silicone wafers, or medical-grade materials) for 24-72 hours under conditions relevant to the research question [46].
- For AFM imaging, immobilize samples using appropriate adhesives such as Cell-Tak, ensuring minimal alteration of native biofilm structure. Optimization of immobilization is critical as division and detachment of cells can occur after multiple divisions if immobilization is insufficient [49].
- For phenotypic assays parallel to AFM, grow biofilms in standardized microbiological media using established protocols such as the Calgary biofilm device or continuous flow systems, ensuring consistency between technical replicates.
Simultaneous AFM-LSCM Imaging Preparation:
- Utilize fully integrated AFM-LSCM systems for correlative imaging. The AFM component should be equipped with quantitative imaging (QI) mode capability, which collects force-distance curves at each pixel, enabling high-resolution mapping of mechanical properties without lateral forces that could displace samples [49].
- For live-cell imaging, maintain physiological conditions throughout experimentation (temperature, pH, nutrient availability) using appropriate environmental chambers.
- For fluorescent tagging, employ genetically encoded fluorescent proteins (e.g., GFP, RFP) targeting specific cellular structures or use vital dyes for monitoring physiological parameters such as reactive oxygen species (e.g., CellROX) [49].
Phenotypic Biofilm Assay Protocols
Crystal Violet Microtiter Assay:
- Grow biofilms in 96-well plates for the desired duration under optimized conditions.
- Carefully remove planktonic cells by washing with phosphate-buffered saline (PBS).
- Fix biofilms with methanol or ethanol for 15 minutes, then stain with 0.1% crystal violet solution for 20 minutes.
- Wash excess stain thoroughly and elute bound crystal violet with 33% acetic acid.
- Quantify biofilm biomass by measuring optical density at 570 nm using a plate reader [46].
- Include appropriate controls (abiotic staining, planktonic cells) and normalize data to cell viability or protein content as needed.
Metabolic Activity Assay:
- Following biofilm growth, add resazurin-based solution (0.015% w/v) to each well.
- Incubate for 30-60 minutes at growth temperature protected from light.
- Measure fluorescence (excitation 530-560 nm, emission 590 nm) to determine metabolic activity.
- Correlate metabolic activity with crystal violet data to distinguish between biofilm biomass and physiological state.
AFM Nanomechanical Characterization
Quantitative Imaging (QI) Mode AFM:
- Use silicon or silicon nitride cantilevers with nominal spring constants of 0.01-0.5 N/m and tip radii of 1-50 nm, depending on required resolution and sample stiffness [49] [48].
- Set imaging parameters to minimize applied force (typically 100-500 pN) while maintaining sufficient signal-to-noise ratio for reliable detection.
- Acquire force-distance curves at each pixel (typically 256 × 256 or 512 × 512 pixels) with a maximum force set point that avoids sample damage.
- For live cell imaging, use scan rates appropriate for the biological process under investigation (e.g., 1.96 line Hz for bacterial cell division studies) [49].
Data Processing and Analysis:
- Convert force-distance curves to Young's modulus values using appropriate contact mechanics models (e.g., Hertz, Sneddon, or Johnson-Kendall-Roberts models).
- Calculate adhesion forces from the minimum force value in retraction curves.
- Determine surface roughness parameters (e.g., RMS roughness) from height images.
- Perform statistical analysis on mechanical properties across multiple samples and biological replicates.
Table 2: Key Nanomechanical Parameters and Their Significance in Biofilm Research
| Parameter | Measurement Principle | Biological Significance | Typical Values for Biofilms | Technical Considerations |
|---|---|---|---|---|
| Young's Modulus (Elasticity) | Slope of force-distance curve during indentation | Matrix stiffness, structural integrity, response to mechanical stress | 0.1-2.0 MPa (varies with EPS composition) | Model selection critical (Hertz, Sneddon); depends on indentation depth |
| Adhesion Force | Minimum force in retraction curve | Cell-surface and cell-cell interactions, polymer adhesion properties | 0.1-5 nN (depends on tip functionalization) | Affected by tip chemistry, loading rate, contact time |
| Surface Roughness | Topographical variation from height images | Biofilm heterogeneity, microcolony formation | RMS: 10-100 nm (depends on growth stage) | Scan size and resolution affect measured values |
| Deformation | Penetration depth at set force | Sample compliance, turgor pressure | 50-500 nm (varies with cellular state) | Must be calibrated for tip geometry |
Data Integration and Analysis Framework
Correlative Data Analysis
The power of this integrated approach lies in the quantitative correlation between nanomechanical properties and phenotypic biofilm characteristics. The following diagram illustrates the relationship between experimental techniques and the parameters they measure:
Statistical Correlation Methodology:
- Perform multivariate analysis to identify relationships between nanomechanical parameters (Young's modulus, adhesion, roughness) and phenotypic measures (biomass, metabolic activity).
- Use machine learning approaches (e.g., principal component analysis, random forests) to identify the most significant parameters distinguishing biofilm phenotypes or treatment responses.
- Implement spatial mapping techniques to correlate local mechanical properties with EPS composition revealed by fluorescent tags or specific molecular probes.
Time-Dependent Analysis:
- For dynamic studies, track changes in nanomechanical properties alongside biofilm development stages from initial attachment to maturation and dispersal.
- Correlate temporal changes in mechanical properties with gene expression data for polysaccharide biosynthesis genes (e.g., icaADBC, alg, psl, pel operons).
- Monitor how antimicrobial treatments alter the mechanical properties of biofilms in relation to traditional viability measures.
Case Study: Pseudomonas aeruginosa Biofilm Analysis
To illustrate the practical application of this correlative approach, consider a study on P. aeruginosa biofilms:
Experimental Design:
- Compare wild-type strains with mutants defective in specific exopolysaccharide biosynthesis (ΔalgD, Δpsl, Δpel).
- Assess biofilm formation using crystal violet assays at 24, 48, and 72 hours.
- Perform AFM nanomechanical mapping on 24-hour biofilms to characterize local variations in stiffness and adhesion.
- Use lectin-based fluorescent probes specific to each exopolysaccharide to correlate spatial distribution with mechanical properties.
Expected Outcomes:
- Psl-deficient mutants would show reduced surface adhesion in early attachment phases, correlating with decreased crystal violet staining.
- Alginate-overproducing strains would exhibit higher matrix stiffness (Young's modulus) and increased resistance to mechanical disruption.
- Pel-producing biofilms would show distinctive adhesion profiles related to their cationic nature and interaction with anionic matrix components.
Essential Research Reagents and Materials
Successful implementation of correlative nanomechanical and phenotypic analysis requires specific research tools and reagents. The following table details essential components of the experimental toolkit:
Table 3: Research Reagent Solutions for Correlative Biofilm Analysis
| Category | Specific Reagents/Materials | Function/Application | Technical Considerations |
|---|---|---|---|
| Biofilm Growth & Staining | Crystal violet (0.1%), Resazurin solution, Calcofluor white, Congo red | Biofilm biomass quantification, metabolic activity assessment, polysaccharide staining | Concentration optimization required for different species; avoid over-fixing for viable AFM samples |
| AFM Consumables | Silicon nitride cantilevers (MLCT, PNP-TR), functionalized tips (lectin-coated), Cell-Tak adhesive | Nanomechanical measurement, specific molecular recognition, sample immobilization | Spring constant calibration critical; functionalization protocols must preserve biological activity |
| Fluorescent Probes | CellROX oxidative stress indicators, SYTO stains, FM lipophilic dyes, GFP/RFP constructs | Viability assessment, ROS detection, membrane integrity, gene expression localization | Photobleaching management; spectral overlap considerations for multiplexing |
| Molecular Biology Tools | Lectin probes (wheat germ agglutinin, concanavalin A), polysaccharide-specific antibodies, gene knockout mutants | EPS component identification, structural characterization, functional validation | Specificity validation required; consider background binding in complex matrices |
| Specialized Media | Mueller-Hinton agar, Tryptic soy broth, defined minimal media with specific carbon sources | Standardized antimicrobial testing, controlled EPS production studies | Composition affects EPS production; consistency critical for reproducibility |
Advanced Applications and Future Directions
High-Content Screening of Anti-biofilm Compounds
The correlative approach described herein enables high-content screening of compounds targeting biofilm matrix integrity. By simultaneously assessing traditional viability metrics (via phenotypic assays) and nanomechanical properties (via AFM), researchers can identify agents that specifically disrupt EPS structure without necessarily killing embedded cells. This is particularly valuable for developing anti-virulence strategies that may exert less selective pressure for resistance development compared to traditional biocides.
Machine Learning-Enhanced Analysis
Recent advances in materials data science provide powerful tools for analyzing complex correlative datasets. Deep learning approaches, particularly U-Net architectures, can achieve pixel-wise classification accuracies exceeding 0.97 for identifying distinct features in microscopy images [50]. These methods enable automated segmentation of biofilm components and quantitative tracking of structural evolution over time. Furthermore, spatiotemporal graph (st-graph) representations can model relationships between particles or microcolonies, capturing cooperative effects and environmental influences that traditional single-particle analyses miss [50].
Clinical Translation and Therapeutic Development
The integration of nanomechanical profiling with phenotypic assays offers new opportunities for clinical diagnostics and therapeutic development. By establishing mechanical signatures associated with treatment-resistant biofilms, clinicians could potentially use AFM-based characterization to guide treatment selection for chronic infections. Additionally, the ability to correlate specific polysaccharide compositions with mechanical properties enables targeted interventions against key matrix components, such as PNAG-degrading dispersin B or alginate lyase treatments to disrupt biofilm integrity [20].
The correlation of AFM nanomechanical data with phenotypic biofilm assays represents a powerful methodological advancement in biofilm research. This integrated approach enables researchers to bridge the gap between macroscopic phenotypic observations and the nanoscale mechanical properties that govern biofilm function and resilience. By providing detailed protocols for sample preparation, data acquisition, and analysis, this technical guide equips researchers with the tools necessary to implement this correlative methodology in their investigation of capsular polysaccharides and other EPS components. As the field continues to evolve, the integration of advanced computational methods and high-throughput screening capabilities will further enhance our ability to decipher the structure-function relationships that underpin biofilm-mediated resistance and persistence in clinical and environmental settings.
Overcoming Analytical Hurdles: Optimizing AFM for Complex Biofilm Systems
Addressing Sample Heterogeneity and Dynamic Properties
Within the field of biofilm research, atomic force microscopy (AFM) nanomechanics provides unparalleled capability to probe the biophysical properties of bacterial capsular polysaccharides at the single-molecule and single-cell level. However, the inherent sample heterogeneity of biological systems and the dynamic nature of polysaccharide polymers present significant methodological challenges that can compromise data interpretation and reproducibility. This technical guide examines the core strategies for addressing these challenges, framed within the broader thesis that understanding the structure-function relationship of capsular polysaccharides is fundamental to developing novel anti-biofilm strategies. We present standardized protocols, quantitative benchmarks, and experimental workflows designed to equip researchers with robust methodologies for extracting meaningful nanomechanical data from complex, variable biological samples, ultimately advancing drug development targeting biofilm-mediated infections.
Quantitative Properties of Capsular Polysaccharides
The biophysical characterization of capsular polysaccharides reveals key parameters that correlate with their biological function, particularly their capacity to inhibit biofilm formation. The table below summarizes quantitative data for a selection of active and inactive polysaccharides, highlighting the critical differentiators.
Table 1: Biophysical Properties of Selected Capsular Polysaccharides
| Polysaccharide | Antibiofilm Activity (Broad-Spectrum) | Molecular Weight (kDa) | Intrinsic Viscosity [η] (dl/g) | Key Electrokinetic Property |
|---|---|---|---|---|
| G2cps | Yes | ~800 | >7 | High density of electrostatic charges |
| Vi | Yes | Not Specified | >7 | Distinct electrokinetic signature |
| MenA | Yes | Not Specified | >7 | Distinct electrokinetic signature |
| PnPS3 | Yes | Not Specified | >7 | Distinct electrokinetic signature |
| PnPS18C | Narrow (S. aureus only) | Not Specified | Intermediate | Intermediate properties |
| PnPS12F | Narrow (E. coli only) | Not Specified | Intermediate | Intermediate properties |
| Inactive Polysaccharides | No | Variable | Systematically low (<7) | Low intrinsic viscosity |
Research indicates that molecular size integrity is a crucial parameter for antibiofilm function. For G2cps, even minor reduction in polysaccharide size via radical oxidation hydrolysis resulted in a complete loss of its antiadhesion properties, underscoring that the conservation of the full-length polymer is critical for its activity [7]. Furthermore, intrinsic viscosity [η], which reflects the volume per mass unit occupied by the polysaccharide in solution and is mediated by its conformation and electrostatic charges, has been shown to be a predictive indicator of activity. All broad-spectrum active macromolecules in a study of 29 different polysaccharides were characterized by a high intrinsic viscosity (>7 dl/g), whereas inactive molecules systematically displayed lower values [7].
Experimental Protocols for AFM Nanomechanics
Probe Functionalization and Calibration
Objective: To ensure consistent and quantitative force measurements by preparing a cantilever with a well-defined tip and known spring constant.
Detailed Methodology:
- Cantilever Selection: Use sharp, non-functionalized silicon or silicon nitride cantilevers for topographical imaging. For force spectroscopy, select cantilevers with appropriate spring constants (typically 0.01-0.1 N/m for living cells [15] [24]).
- Force Constant Calibration: Employ thermal tuning methods to determine the exact spring constant of the cantilever. This involves analyzing the power spectral density of the thermal noise fluctuations of the free cantilever in fluid [51].
- Tip Functionalization (for specific adhesion studies):
- Chemical Functionalization: To measure specific interactions, the AFM tip can be functionalized with relevant molecules (e.g., lectins, antibodies, or host matrix proteins). This is achieved through chemisorption using silane chemistry or physisorption.
- Colloidal Probe Technique: Alternatively, a micron-sized spherical particle (e.g., silica, polystyrene) can be glued to the end of a tipless cantilever. This provides a larger, well-defined contact area for measuring average interaction forces with bacterial cells, reducing the influence of nanoscale heterogeneity [52].
- Probe Characterization: Image the functionalized tip using scanning electron microscopy (SEM) to confirm its geometry and the success of functionalization.
In Situ Nanomechanical Measurement on Live Bacteria
Objective: To quantitatively assess the mechanical properties of the bacterial capsule, such as its stiffness and deformation, under near-physiological conditions.
Detailed Methodology:
- Sample Preparation: Grow bacterial cells to the desired growth phase (e.g., mid-log or stationary phase) to account for physiological heterogeneity. Gently deposit and immobilize the cells on a poly-L-lysine-coated glass slide or a porous membrane filter. Perform all steps in a biologically relevant buffer (e.g., PBS) to maintain cell viability [15] [24].
- AFM Force Curve Acquisition:
- Engage the AFM tip (functionalized or plain) onto the bacterial surface.
- Program the piezoelectric scanner to approach and retract from the cell surface at a constant velocity (typically 0.5-1 µm/s) to minimize hydrodynamic effects.
- Collect a dense grid of force-distance curves (force-volume imaging) over the central region of multiple individual cells to capture spatial heterogeneity. A minimum of 100-500 curves per cell is recommended for statistical significance [15].
- Data Collection Parameters:
- Set-Point Force: Maintain a low applied force (typically 0.2-0.5 nN) to avoid irreversible damage to the cell or its capsule.
- Trigger Threshold: Set a sensitive trigger to detect the initial point of contact with the soft, polymeric capsule.
- Sampling Rate: Use a high sampling rate to capture the full details of the polymer extension and rupture events during retraction.
Data Processing and Analysis of Force Spectroscopy
Objective: To convert raw cantilever deflection data into quantitative nanomechanical parameters and adhesion metrics.
Detailed Methodology:
- Force Curve Conversion: Convert the raw deflection versus displacement data into force-distance curves using the calibrated spring constant and the photodetector sensitivity [51].
- Baseline Correction: Subtract the baseline of the non-contact part of the curve to define zero force.
- Elastic Modulus Fitting: Fit the linear, repulsive region of the approach curve with an appropriate contact mechanics model (e.g., Hertzian, Sneddon, or JKR models) to extract the Young's modulus (stiffness) of the capsule. The choice of model depends on the indenter geometry and material assumptions [15] [24].
- Adhesion Analysis: Analyze the retraction curve to identify adhesion events. Key parameters to extract include:
- Maximum Adhesion Force: The largest force peak in the retraction curve.
- Adhesion Energy (Work of Adhesion): The area under the adhesive part of the retraction curve.
- Rupture Length: The distance at which adhesive bonds break, which can indicate the length of tethered polymers [52].
- Statistical Analysis: Compile data from hundreds of force curves across multiple cells and biological replicates. Present data as mean ± standard deviation and use statistical tests (e.g., ANOVA) to assess the significance of differences between bacterial strains or conditions. This step is critical for addressing sample heterogeneity.
Diagram 1: AFM nanomechanics workflow for live bacteria.
The Scientist's Toolkit: Research Reagent Solutions
The following table catalogues essential materials and their specific functions for conducting robust AFM nanomechanics studies on capsular polysaccharides.
Table 2: Essential Research Reagents and Materials for AFM Nanomechanics
| Research Reagent / Material | Function in Experimental Protocol |
|---|---|
| Silicon Nitride Cantilevers (Sharp Tips) | High-resolution topographical imaging of bacterial cells and their surface structures. |
| Colloidal Probe Cantilebers (Sphere-terminated) | Measuring average interaction forces and nanomechanics with a defined contact geometry, reducing tip variability. |
| Poly-L-Lysine Solution | Coating substrate surfaces to electrostatically immobilize live bacterial cells for AFM measurement. |
| Phosphate Buffered Saline (PBS) | Maintaining physiological conditions and osmotic balance during in-situ force spectroscopy in liquid. |
| Specific Functionalization Ligands (e.g., Lectins, Antibodies) | Grafting onto AFM tips to measure specific receptor-ligand interactions with polysaccharide epitopes. |
| Wild-Type and Isogenic Mutant Bacterial Strains | Comparative studies to directly link specific genetic determinants (e.g., capsule synthesis genes) to nanomechanical properties. |
Visualizing Data Processing and Analysis
The transformation of raw AFM data into quantitative, biologically meaningful information requires a rigorous and multi-step analytical process, as visualized below.
Diagram 2: Data analysis workflow for force spectroscopy.
Atomic Force Microscopy (AFM) has emerged as a pivotal tool in biofilm research, enabling the investigation of capsular polysaccharides at the nanoscale. AFM operates by scanning a sharp tip attached to a flexible cantilever across a sample surface, measuring probe-sample interactions to generate 3D topography images with sub-nanometer resolution [53]. This capability is particularly valuable for studying the mechanical properties of bacterial capsules and their role in biofilm formation. Unlike optical and electron microscopy techniques, AFM can operate in various environments—including ambient air, vacuum, and liquid buffers—making it ideal for examining biological samples in their native states [53]. When studying capsular polysaccharides, researchers can perform in situ nanomechanical measurements of live bacterial cells, providing insights into how these polymer structures influence bacterial adhesion and biofilm development [15] [24].
However, the application of AFM in biofilm research presents two fundamental technical challenges: scan range limitations that constrain the observable areas, and data representativeness concerns regarding the statistical significance of nanomechanical measurements. These challenges are particularly pronounced when studying heterogeneous biofilms and capsular polysaccharides, as these systems exhibit inherent variability across different spatial scales. This technical guide examines these limitations in detail and provides methodologies to overcome them, ensuring reliable and statistically significant results in AFM-based biofilm nanomechanics.
Scan Range Limitations in AFM Biofilm Research
Technical Specifications and Practical Constraints
AFM instruments face inherent trade-offs between scan range, resolution, and imaging speed. Conventional AFM systems typically offer maximum scan ranges of approximately 100μm, with in-plane resolution on the order of several nanometers [53]. This creates a fundamental limitation when studying biofilm systems that may extend across millimeter-scale areas while requiring nanometer-scale resolution to resolve individual polysaccharide chains.
Table 1: AFM Performance Comparison with Other Microscopy Techniques
| Microscopy Technique | Resolution | Typical Image Size | Typical Frame Rate | Main Modality | Environment |
|---|---|---|---|---|---|
| Atomic Force Microscopy (AFM) | 2 nm | 100μm | 0.1 FPS | 3D Topography | vacuum, air, liquid |
| Optical Microscopy | 200 nm | 1000μm | 100 FPS | 2D image | vacuum, air, liquid |
| Scanning Electron Microscopy (SEM) | 10 nm | 1000μm | 20 FPS | 3D image | vacuum |
| Transmission Electron Microscopy (TEM) | 0.2 nm | 100μm | 20 FPS | 2D projection | vacuum |
| Scanning Tunneling Microscopy (STM) | 0.1 nm | 0.5μm | 0.1 FPS | 3D density of states | vacuum, air |
The scan range limitation becomes particularly evident when comparing AFM to other microscopy techniques. While optical microscopy can easily survey millimeter-scale areas to identify regions of interest within heterogeneous biofilms, AFM's narrow field of view may lead to sampling bias if researchers inadvertently select non-representative areas for high-resolution imaging [53]. This limitation is compounded by AFM's relatively slow imaging speed, typically around 0.1 frames per second for conventional systems, making comprehensive large-area mapping time-prohibitive [53].
Impact on Capsular Polysaccharide Research
The restricted scan range of AFM presents specific challenges for studying capsular polysaccharides in biofilm formation:
Limited sampling of bacterial heterogeneity: Individual bacteria within a population exhibit variations in capsular polysaccharide expression, organization, and mechanical properties. AFM's limited scan range may capture only a small subset of this heterogeneity, potentially missing critical patterns in capsule organization that influence biofilm development [15] [24].
Incomplete characterization of biofilm architecture: Biofilms form complex three-dimensional structures with heterogeneous distribution of extracellular polymeric substances. AFM's confined scan range may fail to capture representative features of the overall biofilm architecture, leading to incomplete understanding of how capsular polysaccharides contribute to structural integrity [16].
Challenges in studying cell-cell interactions: The organization of bacterial communities during early biofilm formation occurs over scales that often exceed AFM's typical scan range, making it difficult to capture representative cell-cell interaction events mediated by capsular polysaccharides [15].
Data Representativeness and Statistical Significance
The Statistical Challenge in AFM Nanomechanics
The issue of data representativity extends beyond scan range limitations to encompass the fundamental challenge of deriving statistically significant conclusions from limited AFM measurements. This is particularly critical when studying the nanomechanical properties of capsular polysaccharides, as traditional AFM approaches may collect insufficient data for reliable statistical analysis [54].
The problem arises from multiple factors: the inherent spatial heterogeneity of biological samples, the time-consuming nature of AFM imaging, and the traditional focus on high-resolution imaging of small areas rather than collecting statistically powerful datasets. For capsular polysaccharide research, this means that nanomechanical measurements (such as stiffness, adhesion, and deformation) may not adequately represent the true variability within the bacterial population [15] [24].
High-Speed AFM for Enhanced Statistical Power
Recent advancements in high-speed AFM (HS-AFM) have begun to address these limitations by enabling rapid image acquisition of several frames per second [54]. This technological improvement transforms AFM from a primarily qualitative tool to a quantitative technique capable of collecting large datasets suitable for robust statistical analysis.
Table 2: Statistical Parameters for AFM-Based Quality Control of Nanomechanical Measurements
| Parameter | Description | Application in Capsular Polysaccharide Studies | Recommendation |
|---|---|---|---|
| Sample Size (Number of Measurements) | Number of independent AFM measurements or images collected | Determines ability to detect true differences in polysaccharide mechanical properties | Minimum 30-50 measurements per condition; 200+ for high confidence [54] |
| Measurement Uncertainty | Statistical confidence in measured values | Quantifies reliability of nanomechanical property measurements | Calculate using standard error of the mean for large datasets [54] |
| Area Roughness Parameters (Sa) | Three-dimensional equivalent of Ra roughness | Characterizes surface heterogeneity of bacterial capsules | Preferred over line roughness due to higher statistical significance [54] |
| Line Roughness Parameters (Ra) | Two-dimensional profile roughness | Alternative for rapid assessment of polysaccharide organization | Lower statistical significance; use only for initial screening [54] |
| Intrinsic Viscosity [η] | Measurement of polysaccharide conformation in solution | Correlates with antibiofilm activity; high values (>7 dl/g) indicate activity [16] | Critical parameter for identifying active polysaccharides [16] |
HS-AFM enables researchers to collect hundreds of images rapidly, providing datasets large enough for reliable statistical analysis. For example, one study demonstrated this approach by acquiring over 200 HS-AFM images of silicon carbide fibers, allowing for distinguishing roughness values with high confidence even between very similar samples [54]. This methodology can be directly applied to capsular polysaccharide research, where large datasets enable researchers to account for sample variability and obtain statistically robust measurements of nanomechanical properties.
Experimental Protocols for Representative AFM Nanomechanics
Sample Preparation for Bacterial Capsular Polysaccharides
Proper sample preparation is essential for obtaining representative AFM data on bacterial capsular polysaccharides:
Bacterial Culture and Harvesting:
- Grow Klebsiella pneumoniae (or target bacterial species) under conditions that promote capsule expression [15] [24].
- Harvest bacteria during mid-logarithmic phase by gentle centrifugation (2,000-3,000 × g for 5 minutes).
- Wash cells twice in appropriate buffer (e.g., PBS for physiological conditions) to remove extracellular media components.
Surface Immobilization:
- Functionalize AFM substrates (typically glass or mica) with poly-L-lysine (0.01% w/v) for 30 minutes to enhance bacterial adhesion.
- Apply bacterial suspension to functionalized surface and allow to adhere for 30-60 minutes.
- Gently rinse with appropriate buffer to remove non-adherent cells, maintaining capsule integrity.
Environmental Control:
- For liquid imaging, use appropriate biological buffer that maintains bacterial viability and capsule structure.
- Control temperature if necessary using AFM environmental chamber to maintain physiological conditions.
AFM Imaging Protocol for Nanomechanical Characterization
Standardized imaging protocols ensure consistent and comparable measurements across different samples:
Probe Selection:
- Use sharp silicon nitride tips with nominal spring constants of 0.01-0.1 N/m for contact mode imaging in liquid.
- Select tips with reflective gold coating for optimal laser reflection in optical beam deflection systems.
- Confirm actual spring constant using thermal tuning method before measurements.
Imaging Parameters:
- Set scan size to 1-5μm for individual bacterial imaging, ensuring multiple bacteria are imaged across different sample regions.
- Use scan rates of 0.5-1.0 Hz for conventional AFM, or higher rates (5-10 Hz) for HS-AFM systems.
- Apply minimal loading force (typically 100-500 pN) to avoid compressing or damaging capsular polysaccharides.
Force Spectroscopy Measurements:
- Perform approach-retract cycles at multiple locations on each bacterial cell (minimum 10-15 points per cell).
- Collect curves from at least 30-50 cells per condition to ensure statistical significance.
- Use identical approach and retraction velocities (typically 0.5-1.0 μm/s) across all measurements.
Multi-Scale Imaging Approach
To address scan range limitations while maintaining resolution, implement a multi-scale imaging strategy:
Large-Area Survey Mapping:
- Begin with large scan sizes (50-100μm) at lower resolution to identify representative regions of interest.
- Use optical microscopy integration when available to guide AFM positioning to relevant areas.
Targeted High-Resolution Imaging:
- Select multiple (5-10) representative regions for high-resolution imaging based on survey maps.
- Acquire detailed images (1-5μm) at maximum resolution for nanomechanical analysis.
Correlative Microscopy Integration:
- Combine AFM with optical microscopy to correlate nanomechanical properties with fluorescent labels targeting specific polysaccharide components.
- Use SEM/TEM for ultrastructural characterization of the same samples when feasible.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents for AFM Nanomechanics of Capsular Polysaccharides
| Reagent/Material | Function | Application Example | Considerations |
|---|---|---|---|
| Silicon Nitride AFM Probes | Nanomechanical probing of soft biological samples | Measuring mechanical properties of bacterial capsules [15] [24] | Low spring constants (0.01-0.1 N/m) minimize sample damage |
| Poly-L-Lysine Coated Substrates | Immobilization of bacterial cells | Securing bacteria for AFM imaging without altering capsule structure [15] | Optimize concentration to ensure adhesion while preserving capsule |
| Physiological Buffer Solutions | Maintain native capsule structure during imaging | PBS or other appropriate buffers for bacterial viability [15] | Control ionic strength and pH to match native environment |
| Capsular Polysaccharide Mutants | Control strains for mechanistic studies | Klebsiella pneumoniae mutants with altered capsule organization [15] [24] | Essential for establishing structure-function relationships |
| High Intrinsic Viscosity Polysaccharides | Reference materials for antibiofilm activity | Vi, MenA, MenC polysaccharides with known antibiofilm properties [16] | Intrinsic viscosity >7 dl/g predictive of activity [16] |
Data Processing and Interpretation Framework
Image Processing Considerations
AFM data processing requires careful implementation to avoid introducing artifacts or misinterpretations:
The processing pathway significantly impacts the final data interpretation. For example, applying high-order mathematical functions to subtract background topography can introduce non-physical undulations, potentially leading to incorrect conclusions about surface features [55]. Similarly, inappropriate row alignment can create artificial structures in the final image. Researchers must document all processing steps and validate that processing artifacts do not affect the key features of interest.
Statistical Analysis Protocol
Implement a standardized statistical analysis workflow to ensure data representativeness:
Dataset Sufficiency Assessment:
- Conduct power analysis to determine the minimum number of measurements required for statistical significance.
- Collect additional samples until measurement uncertainty is smaller than the effect size of interest [54].
Variability Quantification:
- Calculate both within-sample and between-sample variability to identify the primary source of measurement variance.
- Use appropriate statistical tests (ANOVA for multiple groups, t-tests for paired comparisons) to assess significance.
Correlation with Biological Activity:
- Correlate nanomechanical measurements with biological outcomes (e.g., biofilm formation capacity).
- For capsular polysaccharides, specifically examine relationships between intrinsic viscosity, nanomechanical properties, and antibiofilm activity [16].
The technical challenges of scan range limitations and data representativeness in AFM nanomechanics of capsular polysaccharides require integrated solutions combining technological advancements, rigorous experimental design, and appropriate data analysis. The implementation of high-speed AFM enables collection of statistically powerful datasets, while standardized protocols ensure consistent and comparable measurements across different laboratories.
Future developments in AFM technology, including increased scan ranges, faster imaging capabilities, and automated multi-scale correlation with other microscopy techniques, will further enhance our ability to study capsular polysaccharides in biofilm formation. Additionally, the integration of machine learning approaches for automated analysis of large AFM datasets promises to extract more sophisticated structure-function relationships from nanomechanical measurements.
By addressing these technical challenges through the methodologies outlined in this guide, researchers can obtain statistically robust, representative data on the nanomechanical properties of capsular polysaccharides, advancing our understanding of their role in biofilm formation and facilitating the development of novel anti-biofilm strategies.
Optimizing Probe-Sample Interactions in Hydrated Conditions
Within the broader scope of Atomic Force Microscopy (AFM) nanomechanics studies of capsular polysaccharides in biofilm research, optimizing probe-sample interactions in hydrated conditions is a critical technical challenge. Biofilms are microbial communities that colonize both biotic and abiotic surfaces, and their formation on indwelling medical devices is a common cause of hospital-acquired infections [15]. The exopolysaccharide capsule has been identified as one of the key bacterial components for biofilm formation, though the underlying biophysical mechanisms remain poorly understood [15]. AFM enables nanomechanical measurements of pathogens like Klebsiella pneumoniae in situ, providing insights into how capsular organization influences bacterial adhesion and biofilm development [15].
This technical guide addresses the specific methodological considerations for investigating soft, hydrated biological samples like bacterial capsules and biofilms using AFM. Unlike hard materials, these samples are soft and compressible, which significantly complicates the interpretation of AFM topography and nanomechanical properties [56]. Under hydrated conditions, the finite force applied by the AFM tip generally results in elastic and/or viscous deformation of the surface, meaning that the measured AFM topography often represents a deformed version of the unperturbed surface [56]. Understanding and controlling these interactions is essential for obtaining accurate, quantitative data on the mechanical properties of capsular polysaccharides and their role in biofilm formation.
Key Challenges in Hydrated AFM Imaging
Interpreting AFM data from soft, hydrated samples requires careful consideration of several technical challenges that can compromise data accuracy.
Sample Deformation and Topographical Errors
A fundamental limitation of AFM is that its operation requires a force between the tip and the sample. For soft materials like hydrated polysaccharide capsules, this force causes deformation, leading to measured topographies that differ substantially from the true surface structure [56]. Finite element modelling (FEM) studies demonstrate that this deformation can cause nanoparticles to appear larger or smaller by a factor of two, depending on tip size and indentation force [56]. Furthermore, a higher spatial resolution in AFM images does not necessarily coincide with a more accurate representation of the sample surface, as increased imaging force often exacerbates deformation artifacts.
Limitations in Spatial Resolution and Mechanical Property Mapping
The spatial resolution in AFM depends fundamentally on the sharpness of the AFM tip [56]. Broader tips cause greater convolution between the tip geometry and sample topography, blurring fine details. This is particularly problematic for the intricate, three-dimensional structure of biofilms. Additionally, the measured elastic modulus in AFM generally deviates from the true elastic modulus of the sample material due to these tip-sample interactions [56]. For accurate mechanical characterization, models must account for the combination of tip-sample geometry and indentation depth, which is especially complex in hydrated environments where capillary forces and solvent layers further influence interactions.
Experimental Protocols for Hydrated Conditions
AFM Force Spectroscopy and Data Analysis
Force spectroscopy is a powerful method for quantifying tip-sample interactions in liquid. In these experiments, the cantilever and tip are moved directly towards the sample until contact is made, then retracted while measuring the interaction [57]. This process is repeated at different locations to map tip-surface interactions or at the same point to build statistical understanding. The data collected in "force curves" can be converted into quantitative measurements of physical properties such as adhesion energy and Young's modulus [57].
Protocol: Basic Force Curve Acquisition in Liquid
- Calibration: Calibrate the cantilever's spring constant and the optical lever sensitivity in fluid prior to sample engagement. Different calibration methods exist, each with advantages and disadvantages that must be considered for the specific experimental setup [57].
- Approach: Position the AFM tip above the region of interest on the hydrated biofilm sample. Initiate the approach cycle, monitoring the cantilever deflection.
- Contact Point Determination: Identify the point of initial contact between the tip and the compliant sample surface. This is a critical step for accurate indentation calculation [56].
- Indentation: Continue piezo movement to apply a defined force, indenting the tip into the soft sample. The resulting cantilever deflection is recorded.
- Retraction: Retract the tip from the surface while recording deflection. Adhesion forces between the tip and sample will manifest as a "pull-off" event in the retraction curve.
- Analysis: Convert raw deflection vs. piezo position data into force vs. tip-sample separation curves. Fit appropriate contact mechanics models (e.g., Hertz, Sneddon, or double-contact models) to the indentation portion of the curve to extract the Young's modulus [56].
Finite Element Modelling for Data Interpretation
Finite Element Modelling (FEM) provides a computational framework to predict how measured AFM topography is affected by mechanical deformation, thereby aiding the interpretation of experimental data [56].
Protocol: FEM Simulation of AFM Indentation
- Define Tip Geometry: Model the AFM tip as a rigid cone with an appropriate opening angle (e.g., θ = 20°) ending in a spherical termination of radius R to resemble common AFM tips [56].
- Define Material Properties: Model the sample (e.g., a bacterial cell with a polysaccharide capsule) as a continuous, homogeneous, and isotropic elastic material. Use literature values for initial estimates (e.g., E = 100 MPa, Poisson ratio ν = 0.3 for biomolecules) [56].
- Set Interaction Parameters: Implement "surface to surface contact" interaction with "hard" normal contact and "rough" tangential contact to simulate non-slip conditions [56].
- Simulate Indentation: Simulate the tip pressing against the elastic sample surface resting on a rigid support. Apply symmetry around the indentation axis to reduce computational complexity.
- Validate with Analytical Models: Compare FEM results with analytical indentation models (Hertz, Sneddon) for simple geometries (e.g., spherical samples) to validate the implementation [56].
- Extract Properties: Use the simulated force-indentation relationship to interpret experimental force curves and derive more accurate mechanical properties, accounting for sample compression effects not considered in simple analytical models.
Quantitative Analysis of Probe-Sample Interactions
Table 1: Comparison of Analytical Models for AFM Indentation Analysis
| Model Name | Applicable Indenter Geometry | Key Formula | Best Use Cases | Limitations |
|---|---|---|---|---|
| Hertz Model [56] | Spherical | F = (4/3) * E* * √(R) * δ^(3/2) |
Small indentations (δ ≪ R) into infinite elastic half-space; validated for spherical tips. | Assumes paraboloid shape; neglects sample compression against substrate; unsuitable for large indentations or conical tips. |
| Sneddon Model [56] | Conical | F = (2/π) * E* * tan(θ) * δ² |
Perfectly conical tips indenting elastic surfaces. | Does not account for the spherical tip termination present on most real AFM tips. |
| Modified Sneddon Model [56] | Conical (adjusted) | F = (2/π) * E* * f(δ) * δ² |
Conical tips indenting spherical samples; provides closer fit to experimental data. | Relies on an empirically fitted function f(δ); remains an approximation. |
| Double Contact Model [56] | Spherical | F_Double = (4/3) * E* * √(R) * δ^(3/2) * (1 + 4 * (R/h)²) |
Thin or confined samples where compression against a rigid substrate (δC) is significant. | More complex than Hertz; requires knowledge or estimation of sample height (h). |
Table 2: Key Parameters for FEM Simulation of AFM on Soft Samples
| Parameter | Description | Typical Values/Range | Impact on Results |
|---|---|---|---|
| Young's Modulus (E) | Intrinsic stiffness of the sample material. | 0.1 - 1000 MPa (for hydrated biological samples) [56] | Directly determines the force-indentation relationship; higher E requires higher force for the same indentation. |
| Poisson's Ratio (ν) | Ratio of lateral to axial strain; measure of compressibility. | ~0.3 - 0.5 (for biomolecules, often assumed to be 0.3) [56] | Affects the apparent modulus E* = E/(1-ν²). |
| Tip Radius (R) | Radius of curvature of the AFM tip's end. | 1 - 60 nm | Larger R causes greater broadening of topographic features and reduces spatial resolution. |
| Opening Angle (θ) | The cone angle of the AFM tip. | 15° - 25° [56] | Influences the contact area and stress distribution during indentation. |
| Indentation Force (F) | The force applied by the tip to the sample. | 10 pN - 100 nN | Higher forces increase sample deformation, leading to underestimated feature heights and overestimated widths. |
Visualization of Workflows and Relationships
Experimental Workflow for Hydrated AFM Nanomechanics
Probe-Sample Interaction Logic
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Research Reagent Solutions for Hydrated AFM Biofilm Studies
| Item Name | Function/Application | Key Considerations |
|---|---|---|
| Functionalized AFM Probes | Modifying tip chemistry to measure specific interactions (e.g., polysaccharide-ligand binding). | Tips can be coated with concanavalin A or other lectins to specifically target capsule polysaccharides. Carboxylate or amine groups enable nonspecific adhesion force measurements. |
| Liquid Imaging Cells (Fluid Cells) | Maintaining sample hydration and enabling buffer exchange during AFM experimentation. | Must be compatible with the specific AFM instrument. Allows for the introduction of drugs, enzymes, or ions to study their effect on capsule mechanics in real-time. |
| Buffer Solutions (e.g., PBS) | Providing a physiologically relevant ionic environment and maintaining biofilm viability. | Ionic concentration affects electrostatic double-layer forces between tip and sample. Must be filtered (0.02 µm) to remove particulate contaminants that can interfere with imaging. |
| Soft Cantilevers | Suitable for sensitive force measurements on compliant biological samples without causing damage. | Low spring constants (typically 0.01 - 0.1 N/m) are essential to minimize indentation force and achieve high signal-to-noise in deflection. |
| Calibration Gratings | Verifying scanner accuracy and tip sharpness in the X, Y, and Z axes. | Used to determine the tip's point spread function, which aids in deconvoluting tip geometry from image features, crucial for accurate size measurement of capsule polymers. |
| Finite Element Modelling Software (e.g., Abaqus) | Computational simulation of tip-sample indentation to derive accurate mechanical properties from force curves. | Enables implementation of complex, multi-layer material models (e.g., a soft capsule on a stiffer cell body) that go beyond simple analytical contact models [56]. |
Integrating AFM with Complementary Imaging and Spectroscopic Techniques
Atomic Force Microscopy (AFM) has established itself as a cornerstone technique in biofilm research, providing unparalleled nanoscale topographical imaging and quantitative nanomechanical property measurements. Its application in studying capsular polysaccharides—key structural components of the biofilm matrix—has revealed critical insights into biofilm mechanics and function. However, a comprehensive understanding of complex microbial systems requires moving beyond standalone AFM applications. The integration of AFM with complementary imaging and spectroscopic techniques creates a synergistic analytical platform that bridges the gap between structural information, chemical composition, and biological activity. This technical guide examines current integrative methodologies, detailing their implementation, applications, and optimization within the specific context of AFM nanomechanics studies of capsular polysaccharides in biofilm research.
AFM Fundamentals in Biofilm Nanomechanics
AFM operates by measuring force interactions between a sharp probe and the sample surface, providing high-resolution topographical data and nanomechanical properties without requiring extensive sample preparation that may alter native structures.
Core AFM Capabilities
- Topographical Analysis: High-resolution 3D surface visualization of biofilm architecture, including extracellular polymeric substance (EPS) distribution and bacterial cell arrangement. [58]
- Particle Analysis: Quantification of the size, shape, and distribution of microbial cells, capsules, and nanoparticles within the biofilm matrix. [58]
- Roughness Analysis: Measurement of surface texture parameters critical for understanding biofilm formation and adhesion properties. [58]
- Force Spectroscopy: Direct measurement of adhesion forces, elastic moduli, and chemical interactions via force-distance curves. [58] [15]
Key AFM Operational Modes in Biofilm Research
Table 1: Primary AFM Operational Modes for Capsular Polysaccharide Characterization
| Mode | Measured Parameters | Biofilm Applications | Resolution Range |
|---|---|---|---|
| Contact Mode | Topography, friction | Surface morphology imaging | Atomic to nanometer |
| Tapping Mode | Topography, phase imaging | Soft capsule visualization | Nanometer scale |
| PeakForce Tapping | Topography, modulus, adhesion | Live bacterial nanomechanics | Nanometer scale |
| Force Volume | Adhesion, elasticity mapping | Polysaccharide mechanical properties | 10-100 nm |
For quantitative studies, particularly those employing High-Speed AFM (HS-AFM) for dynamic process monitoring, robust statistical analysis of large datasets (often exceeding 200 images) is essential to account for sample variability and ensure measurement reliability. [54]
Integrated Technique Methodologies
AFM and Super-Resolution Fluorescence Microscopy
The combination of AFM with super-resolution fluorescence techniques overcomes the diffraction limit of conventional light microscopy, enabling correlated structural and chemical analysis.
Experimental Protocol:
- Sample Preparation: Grow biofilms on glass coverslips suitable for both AFM and optical imaging. For Klebsiella pneumoniae studies, cultivate wild-type and isogenic mutants deficient in capsule or fimbriae production. [15]
- Fluorescence Labeling: Apply fluorescent lectins or antibodies targeting specific capsular polysaccharide epitopes prior to AFM analysis.
- Correlative Imaging:
- Acquire super-resolution images (STORM/STED) to localize specific polysaccharide components.
- Transfer sample to AFM without disturbing biofilm integrity.
- Perform nanomechanical mapping of identical regions using force spectroscopy.
- Data Correlation: Overlay structural and mechanical data with molecular localization maps.
Applications: Direct correlation of polysaccharide distribution with localized mechanical properties; investigation of how specific matrix components contribute to overall biofilm stiffness and adhesion. Nanomechanical measurements of wild-type and mutant Klebsiella pneumoniae have demonstrated that capsule organization significantly influences bacterial adhesion and biofilm formation. [15]
AFM and Raman Spectroscopy
This integration provides simultaneous topographical, mechanical, and chemical information from the same biofilm location without requiring external labels.
Experimental Protocol:
- Integrated System Setup: Utilize combined AFM-Raman systems with reflective substrates (e.g., gold-coated glass) to enhance Raman signal.
- Coordinate Registration: Establish spatial correlation between AFM tip position and Raman laser focus.
- Simultaneous Data Acquisition:
- Engage AFM tip in tapping mode to minimize sample disturbance.
- Acquire Raman spectra (typically 400-1800 cm⁻¹ range) with integration times of 0.5-2 seconds per spectrum.
- Hyperspectral Mapping: Collect Raman spectra at predetermined grid points to construct chemical maps correlated with topographical features.
Applications: In situ characterization of polysaccharide chemical composition and conformation; monitoring of metabolic responses to environmental stressors; identification of chemical heterogeneities within biofilm subregions. Raman spectroscopy offers molecular-level fingerprints that elucidate the chemical compositions of microbial communities. [59]
AFM and Scanning Electron Microscopy (SEM)
Correlative AFM-SEM provides comprehensive topological and ultrastructural analysis, with AFM supplying quantitative height data and nanomechanics to complement SEM's high-resolution surface imaging.
Experimental Protocol:
- Sample Preparation:
- For SEM: Fix biofilms with glutaraldehyde (2.5% in buffer), dehydrate in ethanol series, and critical point dry.
- Optional: Sputter-coat with thin (2-5 nm) conductive layer for high-resolution SEM while preserving AFM capability.
- Sequential Imaging:
- Acquire SEM images of regions of interest at various magnifications.
- Transfer to AFM for nanomechanical characterization of identical locations.
- Data Integration: Register AFM and SEM images using distinctive topological features as landmarks.
Applications: Detailed analysis of capsule-bacteria interactions; visualization of EPS fibril networks; correlation of surface morphology with mechanical properties in genetically modified strains. SEM provides high-resolution surface images of biofilms, revealing structural organization, cellular arrangement, and extracellular matrix features. [60]
AFM and Microfluidics
Microfluidic platforms enable real-time observation of biofilm development under controlled hydrodynamic conditions, with integrated AFM providing intermittent nanomechanical characterization.
Experimental Protocol:
- Microfluidic Device Fabrication: Create polydimethylsiloxane (PDMS)-based flow cells with optically clear viewing regions and appropriate channel geometry. [59]
- Experimental Setup:
- Mount microfluidic device on compatible AFM stage.
- Inoculate with bacterial suspension and initiate flow of growth medium.
- Allow biofilm development under controlled shear conditions.
- Time-Lapse Analysis:
- Periodically pause flow for AFM nanomechanical measurements.
- Resume flow between measurements to monitor dynamic responses.
Applications: Investigation of capsule mechanical adaptation to fluid shear stress; real-time monitoring of biofilm development; assessment of antibiofilm agent efficacy under physiologically relevant conditions. Microfluidic chips and flow cells enable detailed, rapid, and precise analyses of microbial communities and their interactions with contaminants. [59]
Research Reagent Solutions
Table 2: Essential Research Reagents for AFM-Based Biofilm Nanomechanics
| Reagent/Category | Specific Examples | Function in Experimental Workflow |
|---|---|---|
| Bacterial Strains | Wild-type & isogenic mutants of Klebsiella pneumoniae | Comparative studies of capsule & fimbriae functions [15] |
| Growth Media | Lysogeny Broth (LB), minimal media with varying osmolarity | Culture maintenance & stress condition studies [23] |
| Fluorescent Labels | FITC-conjugated lectins, polysaccharide-specific antibodies | Specific polysaccharide visualization in correlative microscopy |
| Fixation Reagents | Glutaraldehyde, paraformaldehyde | Sample preservation for SEM & structural studies [60] |
| AFM Probes | Silicon nitride tips, sharp silicon tips | Nanomechanical property measurement & high-resolution imaging [58] |
| Microfluidic Components | PDMS, perfusion chambers | Controlled hydrodynamic condition creation [59] |
Experimental Workflow and Data Integration
The successful integration of multiple techniques requires careful experimental design and data correlation strategies. The following workflow diagram illustrates a standardized approach for correlated AFM and complementary imaging in biofilm studies:
Integrated Experimental Workflow for AFM and Complementary Techniques
Data Correlation Methodology
Effective integration requires systematic registration of multi-modal datasets:
- Landmark-Based Registration: Identify distinctive topological features visible across multiple techniques for spatial alignment.
- Coordinate Transformation: Develop transformation matrices to map coordinates between different imaging systems.
- Multi-Channel Data Fusion: Combine data streams into unified representations using specialized software platforms.
Technical Challenges and Optimization Strategies
Sample Preparation Considerations
Maintaining biofilm structural integrity while meeting requirements of multiple techniques presents significant challenges:
- Fixation Artifacts: Chemical fixation for SEM can alter nanomechanical properties, necessitating careful comparison with live samples. [60]
- Substrate Compatibility: Must accommodate both AFM scanning and optical transparency for correlative microscopy.
- Hydration State: Air-drying for SEM collapses hydrogel-like capsules, while AFM of hydrated samples requires careful fluid control.
Spatial and Temporal Resolution Matching
Mismatched resolution and sampling timescales between techniques can complicate data interpretation:
- Resolution Bridging: AFM provides nanometer spatial resolution, while optical techniques typically offer 200-300 nm resolution at best.
- Temporal Synchronization: AFM image acquisition may require minutes, while dynamic biofilm processes occur in real-time.
Future Perspectives
The integration of AFM with complementary techniques continues to evolve with several promising directions:
- Cryo-Based Correlative Methods: Cryo-electron microscopy combined with cryo-AFM preserves native biofilm structure without chemical fixation artifacts. [59]
- AI-Enhanced Data Integration: Machine learning algorithms rapidly correlate multi-modal datasets, identifying non-intuitive relationships between chemical, structural, and mechanical properties. [59]
- High-Speed AFM Developments: HS-AFM enables statistical analysis of large image datasets (200+ images) for reliable quantification of biofilm properties. [54]
The integration of AFM with complementary imaging and spectroscopic techniques represents a powerful paradigm for advancing biofilm research, particularly in understanding the structure-function relationship of capsular polysaccharides. While technical challenges remain in sample preparation, data correlation, and platform integration, the synergistic insights gained from these multi-modal approaches far outweigh these limitations. As these methodologies continue to mature and incorporate emerging computational and instrumental advances, they will undoubtedly uncover new fundamental principles of biofilm mechanics and enable innovative therapeutic strategies for combating biofilm-associated infections.
Best Practices for Sample Preparation and Data Interpretation
Atomic force microscopy (AFM) has emerged as a powerful, multifunctional platform for probing the nanomechanical properties of capsular polysaccharides and their critical role in biofilm architecture and resilience. This technique provides unique capabilities for quantifying the mechanical forces that govern bacterial adhesion, biofilm assembly, and the functional integrity of extracellular polymeric substances under native conditions. The application of AFM nanomechanics in biofilm research has revealed fundamental structure-function relationships in microbial systems, offering unprecedented insight into microbial colonization strategies and potential interventions for biofilm-associated infections [61]. This technical guide outlines current best practices for sample preparation, AFM operation, and data interpretation specifically framed for researchers investigating the mechanical properties of capsular polysaccharides within biofilm systems.
Sample Preparation Methodologies
Substrate Selection and Functionalization
Proper substrate preparation is foundational for reliable AFM analysis of biofilm polysaccharides. The ideal substrate provides secure immobilization while minimizing alteration of native microbiological properties.
- Adhesion-Promoting Surfaces: Use freshly cleaved mica or silicon wafers treated with poly-L-lysine or trimethoxysilyl-propyl-diethylenetriamine to enhance bacterial adhesion through electrostatic interactions [61].
- Chemically Defined Surfaces: For studies investigating surface-specific effects on biofilm formation, employ functionalized surfaces such as PFOTS-treated glass coverslips to create controlled hydrophobic/hydrophilic interfaces [42].
- Combinatorial Approaches: Utilize gradient-structured surfaces to systematically study how varying surface properties influence attachment dynamics and community structure in a single experiment [42].
Bacterial Immobilization Techniques
Secure immobilization of microbial cells is essential for AFM imaging and force measurements but must preserve cellular viability and native mechanical properties.
Table 1: Bacterial Immobilization Methods for AFM Analysis
| Method Type | Specific Approach | Best For | Considerations |
|---|---|---|---|
| Mechanical | Porous membranes/agar with pore diameters similar to cell size [61] | Single-cell analysis | Sporadic immobilization; reduced reproducibility |
| Mechanical | PDMS microstructures with customizable dimensions (1.5-6 µm wide, 1-4 µm depth) [61] | Spherical microorganisms | High immobilization security; accommodates various cell sizes |
| Chemical | Poly-L-lysine, carboxyl group cross-linking [61] | High-resolution imaging | Potential impact on nanomechanical properties |
| Physiological | Addition of divalent cations (Mg²⁺, Ca²⁺) and glucose [61] | Viability-critical studies | Maintains cellular viability while promoting attachment |
Sample Preservation and Hydration Control
Maintaining native hydration conditions is critical for preserving the structural and mechanical integrity of capsular polysaccharides during AFM analysis.
- Physiological Conditions: Perform imaging and force measurements in liquid cells using appropriate buffers (e.g., PBS) to maintain polysaccharide conformation and biological activity [61].
- Minimal Fixation: Avoid chemical fixatives when possible; if necessary, use gentle fixation methods such as low concentrations of glutaraldehyde (0.1-0.5%) to stabilize structures without significantly altering mechanical properties [61].
- Controlled Drying: For high-resolution topographical imaging of specific structures like flagella, use gentle rinsing and air-drying protocols [42].
AFM Operational Modes and Parameter Optimization
Imaging Modalities for Polysaccharide Characterization
Selecting the appropriate AFM imaging mode is essential for resolving the delicate structures of capsular polysaccharides without inducing damage or artifacts.
- Tapping Mode: The preferred method for imaging soft biological samples like polysaccharides and intact biofilms. This intermittent contact mode reduces friction and drag forces that could damage delicate extracellular structures [61]. Operational parameters should be optimized with free oscillation amplitudes of 10-100 nm and setpoint ratios between 0.7-0.9 to balance image quality and sample preservation.
- Phase Imaging: Capture simultaneously with topographical data to distinguish materials on heterogeneous surfaces. Phase angle changes provide qualitative information about mechanical properties and tip-sample contact area, helping differentiate polysaccharide-rich regions from cellular components [61].
- High-Speed AFM (HS-AFM): Employ for dynamic studies of biofilm formation and polysaccharide rearrangement. HS-AFM enables collection of statistically powerful datasets with several frames per second, ideal for capturing temporal changes in polysaccharide organization during biofilm development [54].
Force Spectroscopy for Nanomechanical Characterization
AFM force spectroscopy provides direct quantification of the mechanical properties relevant to polysaccharide function in biofilms.
- Adhesion Force Mapping: Use functionalized tips (with specific chemical groups or microbial cells) to measure binding forces between polysaccharides and surfaces or other cells. These measurements reveal the nanoscale forces governing initial bacterial attachment and biofilm cohesion [61].
- Nanoindentation Experiments: Quantify mechanical properties including elastic modulus, turgor pressure, and viscoelasticity of individual cells and biofilm matrices. The Hertz model with parabolic tip approximation is most commonly applied for analyzing force-indentation data [61].
- Single-Molecule Force Spectroscopy: Study polysaccharide folding pathways and polymer mechanics by attaching individual polysaccharide chains to AFM tips and monitoring force-extension relationships during stretching and relaxation cycles.
Table 2: Key Parameters for AFM Nanomechanics of Biofilm Polysaccharides
| Measurement Type | Critical Parameters | Typical Values | Data Interpretation Models |
|---|---|---|---|
| Topographical Imaging | Scanning rate: 0.5-2 HzSetpoint: 0.5-1.0 nNResolution: 512×512 pixels | Roughness (Sa): 34-53 nm (SiC fibres) [54] | Statistical analysis of large datasets (>200 images) [54] |
| Adhesion Forces | Approach/retract speed: 0.5-2 µm/sContact time: 0.1-1 sTrigger threshold: 10-100 pN | Capsular polysaccharide interactions: 0.1-5 nN [61] | Adhesion frequency histograms,Bell-Evans model for kinetics |
| Elastic Modulus | Indentation depth: <10% sample heightLoading rate: 0.1-10 nN/sCantilever spring constant: 0.01-0.5 N/m | Bacterial cells: 0.5-5 MPa [61]EPS matrix: 0.1-1 kPa [61] | Hertz model (parabolic tip),Sneddon model (conical tip) |
| Polymer Mechanics | Stretching velocity: 0.1-10 µm/sSampling rate: 10-100 kHz | Polysaccharide contour length: 100-1000 nm [61] | Worm-like Chain model,Freely Jointed Chain model |
Data Interpretation and Analytical Frameworks
Quantitative Analysis of Nanomechanical Data
Proper interpretation of AFM data requires robust theoretical frameworks and statistical approaches to extract meaningful biological insights.
Theoretical Models for Mechanical Properties: Apply the Hertz model for analyzing nanoindentation data, which describes the elastic deformation of two perfectly homogeneous smooth bodies touching under load. The model is expressed as:
( F = \frac{4}{3} \frac{E}{1-\nu^2} \sqrt{R} \delta^{3/2} )
where F is the force on the cantilever, E is the Young's modulus, ν is the Poisson ratio, R is the tip radius, and δ is the indentation depth [61].
Large-Area Statistical Analysis: Implement automated analysis pipelines for large AFM datasets (>200 images) to achieve statistical significance in roughness measurements and mechanical property mapping. Small measurement uncertainties from large datasets enable distinction of even highly similar samples [54].
- Machine Learning-Enhanced Analysis: Utilize machine learning algorithms for automated segmentation, classification, and feature detection in AFM images. These approaches efficiently extract parameters such as cell count, confluency, cell shape, and orientation across extensive areas [42].
Correlation of Nanomechanics with Biochemical Properties
Integrating AFM data with biochemical and structural information provides comprehensive understanding of polysaccharide function in biofilms.
- Structure-Mechanics-Function Relationships: Correlate polysaccharide structural features (molecular weight, charge density, conformation) with measured mechanical properties and antibiofilm activity. Studies show that polysaccharides with high intrinsic viscosity (>7 dl/g) and specific electrokinetic signatures often exhibit broad-spectrum antibiofilm activity [7] [16].
- Size-Activity Correlations: Recognize that polysaccharide size integrity is critical for antiadhesion properties. Even minor reduction of polysaccharide size through fragmentation can result in complete loss of antibiofilm activity, highlighting the importance of polymer length in functional mechanisms [7] [16].
- Multiparametric Analysis: Combine topographical, mechanical, and chemical data to build comprehensive models of biofilm organization and function. Advanced AFM platforms can simultaneously map topographical features, adhesion forces, elastic modulus, and chemical composition [61].
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Research Reagent Solutions for AFM Biofilm Studies
| Reagent/Material | Function | Application Notes |
|---|---|---|
| PFOTS-treated glass | Creates controlled hydrophobic surfaces | Promotes specific bacterial attachment patterns; reveals preferred cellular orientation [42] |
| Polydimethylsiloxane (PDMS) microstructures | Mechanical cell entrapment | Customizable dimensions (1.5-6 µm width, 1-4 µm depth) for various cell sizes [61] |
| Poly-L-lysine | Chemical immobilization agent | Provides strong adhesion but may affect nanomechanical properties [61] |
| Divalent cations (Mg²⁺, Ca²⁺) | Physiological immobilization | Maintains cellular viability while promoting attachment; often used with glucose [61] |
| Capsular polysaccharides (G2cps, Vi, MenA) | Antiadhesion agents | High intrinsic viscosity (>7 dl/g) correlates with broad-spectrum antibiofilm activity [7] [16] |
| High molecular weight polysaccharides | Biofilm matrix components | Size integrity critical for function; molecular weights ~800 kDa maintain antiadhesion properties [7] [16] |
Advanced Applications and Future Directions
Large-Area AFM and Automated Imaging
Traditional AFM's limited scan area has been a significant constraint in biofilm research, but recent advancements have overcome this limitation.
- Automated Large-Area AFM: Implement systems capable of capturing high-resolution images over millimeter-scale areas, enabling comprehensive analysis of biofilm heterogeneity and organization. These systems use automated scanning procedures with minimal user intervention [42].
- Image Stitching Algorithms: Employ sophisticated stitching algorithms that perform well with minimal matching features between images. Limited overlap between scans maximizes acquisition speed while producing seamless, high-resolution composites that capture spatial complexity [42].
- Machine Learning Integration: Utilize AI-driven models for optimal scanning site selection, reducing human intervention and accelerating data acquisition. These systems can autonomously identify regions of interest based on predefined criteria [42].
Correlative Microscopy and Multimodal Approaches
No single technique can fully characterize the complexity of biofilm systems, making correlative approaches essential.
- Integration with Optical Microscopy: Combine AFM with fluorescence microscopy to correlate mechanical properties with specific cellular components or metabolic states.
- Complementary Spectroscopy: Augment AFM data with Raman spectroscopy or Fourier transform infrared spectroscopy to obtain chemical information corresponding to topographical and mechanical features [42].
- SEM/AFM Correlation: Although SEM requires sample dehydration and metal coating that can alter structures, it can provide valuable complementary information when carefully correlated with AFM data [42].
The continued advancement of AFM technologies, particularly in automation, large-area scanning, and machine-learning enhanced analysis, is transforming our ability to quantitatively characterize the nanomechanical properties of capsular polysaccharides in biofilms. By following these best practices in sample preparation, instrument operation, and data interpretation, researchers can generate robust, statistically significant insights into the mechanical world of microbial communities, accelerating the development of novel anti-biofilm strategies and materials.
Beyond Single-Species Analysis: Validation and Broader Implications
Validating AFM Findings with Microtiter Plate and Flow-Cell Assays
Atomic Force Microscopy (AFM) has emerged as a powerful tool in microbiology for probing the nanomechanical properties and ultrastructure of microbial cell surfaces under physiological conditions. Unlike electron microscopy techniques, AFM enables researchers to image living microbial cells in buffer solution at molecular resolution, while simultaneously measuring their physical properties. This capability is particularly valuable for studying capsular polysaccharides—key virulence factors that protect bacterial pathogens from environmental stresses and host immune responses. However, AFM findings require validation through established biological assays to confirm their functional significance in biofilm formation and development.
The integration of AFM with conventional biofilm assays creates a powerful correlative approach that links nanoscale structural and mechanical observations with population-level phenotypic outcomes. This technical guide provides detailed methodologies for validating AFM-based nanomechanics research on capsular polysaccharides using two established biofilm assessment platforms: static microtiter plate assays and dynamic flow-cell systems. By implementing these validation strategies, researchers can bridge the gap between single-molecule measurements and community-level biofilm behaviors, providing compelling evidence for the functional role of specific biophysical properties in bacterial pathogenesis and antimicrobial resistance.
Core Validation Methodologies
Atomic Force Microscopy for Capsular Polysaccharide Characterization
AFM provides unprecedented capability for visualizing capsular polysaccharides and measuring their nanomechanical properties on live bacterial cells. The technique operates by sensing interaction forces between a sharp tip and the sample surface, generating high-resolution three-dimensional images of cell surface architecture without requiring staining, labeling, or fixation [62]. For capsular polysaccharide research, AFM can directly image the rough morphology decorated with nanoscale waves that characterize extracellular polysaccharides on probiotic bacteria like Lactobacillus rhamnosus GG [62].
Key AFM Methodologies:
- Topographic Imaging: The AFM tip follows cell contours in solution to generate 3D images of capsular architecture with near-molecular resolution, typically using contact or tapping mode in liquid environment.
- Single-Molecule Force Spectroscopy (SMFS): The tip is functionalized with specific biomolecules (e.g., lectins) and approached/retracted from the sample to measure interaction forces as a function of distance, quantifying the localization, binding strength, and mechanics of single polysaccharide chains [62].
- Single-Cell Force Spectroscopy (SCFS): A variation where the tip is replaced by a living cell to probe single-cell adhesion forces, revealing how capsular polysaccharides mediate bacterium-substratum interactions [62].
Recent studies have employed time-resolved AFM imaging to investigate antimicrobial peptide interactions with bacterial capsules, revealing localized defects in cell walls while leaving capsular polysaccharides unchanged [63]. This demonstrates AFM's unique capability to simultaneously track structural changes and mechanical properties during therapeutic interventions.
Microtiter Plate Biofilm Assay
The microtiter plate assay provides a high-throughput method for quantifying biofilm formation capacity, enabling researchers to correlate AFM-measured polysaccharide properties with adhesion potential across multiple bacterial strains or conditions.
Protocol:
- Culture Preparation: Grow bacterial strains in appropriate media (e.g., lysogeny broth) with shaking (220 rpm) for 24 hours at 37°C [64].
- Normalization: Dilute overnight cultures to an optical density (OD600) of 0.5 in biofilm media (e.g., tryptic soy broth with 0.5% glucose) [64].
- Biofilm Growth: Transfer 1-2 mL aliquots to tissue culture-treated 6-well plates or 100-200 μL to 96-well plates. Incubate statically for 24 hours at 37°C [64].
- Staining: Carefully remove supernatant and wash biofilms with 1 mL phosphate-buffered saline (PBS) to remove non-adherent cells. Add 0.1% crystal violet solution (1 mL for 6-well plates, 100-200 μL for 96-well plates) and incubate for 15 minutes [64].
- Destaining and Quantification: Remove crystal violet and wash gently with PBS. Add 30% acetic acid (1 mL for 6-well plates, 100-200 μL for 96-well plates) and incubate for 15 minutes to dissolve the stain. Transfer the solution to a new 96-well plate and measure optical density at 550 nm using a microplate reader [64].
- Controls and Normalization: Include control wells with sterile biofilm media processed identically to sample wells. Subtract control OD550 values from sample values to account for dye binding to plastic surfaces.
Table 1: Quantitative Data from Microtiter Plate Biofilm Assays in K. pneumoniae Studies
| Strain Characteristic | Biofilm Formation (OD550) | Mucoviscosity (%) | Correlation with Capsule |
|---|---|---|---|
| Strong biofilm formers | >5.0 | Variable | Positive or negative |
| Moderate biofilm formers | 2.0-5.0 | Variable | Strain-dependent |
| Weak biofilm formers | 1.0-2.0 | Variable | Strain-dependent |
| Very weak biofilm formers | <1.0 | Variable | Often negative |
This protocol can be modified to assess the antibiofilm activity of purified capsular polysaccharides by including them in the biofilm media at specific concentrations (e.g., 100 μg/mL) [16]. The percent inhibition can be calculated by comparing biofilm formation in treated versus untreated controls.
Flow-Cell Biofilm Assay
Flow-cell systems provide a more physiologically relevant environment for biofilm studies by enabling continuous nutrient delivery and waste removal, mimicking natural conditions where biofilms develop under hydrodynamic forces.
Protocol:
- System Setup: Assemble flow-cell chambers with appropriate surface properties (typically glass or plastic coverslips). Sterilize the system before inoculation [16].
- Inoculation: Introduce bacterial suspensions (OD600 ≈ 0.5) into the flow chambers and allow for initial adhesion phase (typically 1-2 hours without flow).
- Medium perfusion: Initiate continuous flow of dilute nutrient medium (e.g., 1/10 or 1/100 strength tryptic soy broth) at controlled rates (typically 0.1-0.5 mm/s) using a peristaltic pump for 24-72 hours [16].
- Staining and Visualization: Introduce fluorescent stains (e.g., SYTO9 for cells, calcofluor white for polysaccharides) into the flow system. Alternatively, fix biofilms with paraformaldehyde for post-staining.
- Imaging and Analysis: Examine biofilms using confocal laser scanning microscopy or equivalent techniques. Acquire Z-stacks at multiple positions for quantitative analysis of biofilm thickness, biovolume, and spatial organization.
- Image Analysis: Process confocal data using software such as ImageJ (with BiofilmQ plugin) or COMSTAT to extract quantitative parameters about biofilm architecture.
For evaluating antibiofilm polysaccharides, these compounds can be introduced during the adhesion phase or after biofilm establishment to assess preventive or disruptive activity, respectively [16].
Table 2: Comparative Analysis of Biofilm Assay Platforms
| Parameter | Microtiter Plate Assay | Flow-Cell Assay |
|---|---|---|
| Throughput | High (multiple conditions) | Low (limited chambers) |
| Physiological relevance | Static conditions | Dynamic fluid flow |
| Biofilm architecture | Limited spatial organization | Complex 3D structures |
| Sample recovery | Destructive (endpoint) | Non-destructive (time-course) |
| Key readouts | Total biomass (OD550) | Thickness, biovolume, spatial distribution |
| Resource requirements | Low (standard lab equipment) | High (specialized flow system, confocal microscope) |
Experimental Workflow Integration
The power of combining AFM with biofilm assays lies in the complementary data each technique provides. The following workflow diagram illustrates how these methods integrate to validate the role of capsular polysaccharides in biofilm formation:
Research Reagent Solutions
Table 3: Essential Research Reagents for AFM and Biofilm Studies
| Reagent/Category | Specific Examples | Function/Application |
|---|---|---|
| AFM Consumables | Silicon nitride tips, cantilevers | Surface probing and force measurement |
| Biofilm Stains | Crystal violet, SYTO9, calcofluor white | Biomass quantification and matrix visualization |
| Polysaccharide Characterization Tools | Lectin-functionalized tips, HPSEC-LS, NMR | Single-molecule analysis and structural characterization |
| Growth Media | Tryptic soy broth, lysogeny broth, OptiMEM | Biofilm promotion under controlled conditions |
| Surface Materials | Glass coverslips, tissue culture plastic | Substrata for biofilm growth |
| Fixation Reagents | Paraformaldehyde, glutaraldehyde | Sample preservation for imaging |
Data Interpretation and Correlation Strategies
Successful validation of AFM findings requires strategic correlation of nanomechanical data with biofilm phenotypic outcomes. Key correlation strategies include:
Linking Biophysical Properties to Anti-biofilm Activity: Studies have demonstrated that antibiofilm capsular polysaccharides share distinct biophysical properties, including high intrinsic viscosity (>7 dL/g) and specific electrokinetic signatures [16]. When AFM identifies polysaccharides with these properties, researchers can hypothesize antibiofilm function and test this using microtiter plate and flow-cell assays.
Connecting Nanoscale Architecture to Biofilm Phenotype: AFM imaging can reveal differences in polysaccharide organization between strong and weak biofilm-forming strains. For example, strains with exposed peptidoglycan layers (detected using LysM-functionalized tips) may show enhanced adhesion in biofilm assays [62].
Relating Single-Molecule Mechanics to Community Behavior: SMFS measurements of polysaccharide adhesion forces can be correlated with biofilm stability under flow conditions. Higher unbinding forces for specific polysaccharide-ligand interactions may predict enhanced biofilm resilience in flow-cell systems.
The integration of these methodologies creates a robust framework for validating the functional significance of AFM-based observations, strengthening conclusions about structure-function relationships in bacterial capsules and their role in biofilm development. This multimodal approach provides compelling evidence that bridges single-molecule measurements with population-level phenotypes, offering insights that no single technique could provide independently.
Comparative Nanomechanics Across ESKAPE Pathogens
The ESKAPE pathogens—Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species—represent a group of clinically critical bacteria renowned for their ability to "escape" the biocidal effects of conventional antibiotics [65]. These pathogens are leading causes of multidrug-resistant (MDR) nosocomial infections, contributing significantly to the global antimicrobial resistance (AMR) burden that is projected to cause 10 million deaths annually by 2050 [65].
Within the context of biofilm research, the capsular polysaccharide (CPS) constitutes a major virulence determinant for many bacterial pathogens, forming a protective layer that surrounds the cell and is critically involved in surface adhesion, biofilm formation, and immune evasion [15]. The nanomechanical properties of this capsule, and its interaction with other surface structures, fundamentally influence the initial stages of bacterial colonization [66] [15]. Atomic force microscopy (AFM) has emerged as a powerful tool for quantifying these nanomechanical properties under physiologically relevant conditions, providing unique insights into the biophysical mechanisms of pathogenicity that complement molecular and genetic studies [66]. This technical guide synthesizes current AFM-based nanomechanics research to compare the mechanical properties and adhesion mechanisms across ESKAPE pathogens, with a specific focus on the role of capsular polysaccharides in biofilm development.
Atomic Force Microscopy in Pathogen Nanomechanics
Fundamental Principles of AFM
Atomic force microscopy operates by physically scanning a cantilever-mounted tip across a sample surface. The interaction forces between the tip and the sample cause cantilever deflections, which are monitored via a laser spot reflected from the top of the cantilever onto a photodetector [66]. These deflections provide feedback to piezoelectric positioners that maintain a constant force, enabling the reconstruction of topographical images with sub-nanometer resolution. More importantly for nanomechanics, AFM can quantify adhesion forces, elasticity (Young's modulus), and stiffness through force-distance curve measurements, wherein the tip is approached toward, makes contact with, and is retracted from the sample surface [66].
A key advantage of AFM in microbiology is its ability to probe live cells in aqueous environments, preserving native structures and functions. This allows for real-time observation of dynamic processes and mechanical responses of pathogens [66]. However, researchers must be aware of inherent limitations, including potential tip-induced sample deformation, a relatively small field of view, and sensitivity to environmental noise [66]. Furthermore, a critical consideration is tip effects, where the scanning tip itself can disrupt molecular interactions, even in strongly binding systems like streptavidin-biotin complexes [67].
Essential AFM Methodologies for Bacterial Nanomechanics
- Sample Immobilization: Gentle immobilization of live bacterial cells is crucial. Common methods include:
- Physical Adsorption: Depositing cells onto freshly cleaved mica or glass substrates, sometimes functionalized with poly-L-lysine or concanavalin A to enhance adhesion.
- Porous Membrane Trapping: Filtering a bacterial suspension onto a porous polycarbonate membrane, which physically traps cells for measurement [66].
- Single-Cell Force Spectroscopy (SCFS): This technique directly measures the interaction forces between a single bacterial cell (attached to the AFM tip via a colloidal probe or chemical linker) and a substrate or host receptor. SCFS quantifies adhesion forces and their spatial distribution [66].
- Chemical Force Microscopy (CFM): The AFM tip is functionalized with specific molecular ligands (e.g., carbohydrates, host proteins). This allows mapping of receptor-ligand interactions on the bacterial surface, quantifying binding forces and kinetics [66].
- High-Speed AFM (HS-AFM): HS-AFM dramatically increases imaging rates from minutes to seconds per frame, enabling the visualization of dynamic processes like the activity of surface proteins or the real-time interaction of pathogens with surfaces [67].
Comparative Nanomechanics of ESKAPE Pathogens
The following section and table summarize key nanomechanical findings for ESKAPE pathogens, with an emphasis on capsule-mediated properties.
Table 1: Comparative Nanomechanics of Select ESKAPE Pathogens
| Pathogen | Key Surface Structure | Nanomechanical Property/Adhesion Force | Influence on Biofilm & Notes |
|---|---|---|---|
| Klebsiella pneumoniae | Capsular Polysaccharide (CPS) | Young's Modulus: Reduced in highly encapsulated strains; Organization influenced by type 3 fimbriae [15]. | Softer, organized capsule promotes initial adhesion and biofilm formation; fimbriae compact the capsule, facilitating contact with surfaces [15]. |
| Pseudomonas aeruginosa | Type IV Pili | Adhesion Force: ~0.2 - 2 nN per pilus; exhibits spring-like properties with high extensibility [66]. | Pili mediate "twitching motility" essential for surface exploration and microcolony formation; dynamic attachment under fluid flow [66]. |
| Staphylococcus aureus | Surface proteins (e.g., FnBPs), Biofilm Matrix | High cellular stiffness in MRSA; strong, multifactorial adhesion mediated by multiple surface molecules [66]. | Robust biofilm formation on abiotic surfaces (e.g., medical devices); adhesion is reinforced by a combination of specific and non-specific interactions [66]. |
| Acinetobacter baumannii | CPS, Outer Membrane Proteins | Young's Modulus: Varies with capsule thickness; adhesive properties altered in MDR strains [65]. | Capsule confers a "slippery" or "sticky" phenotype, directly impacting initial surface attachment and persistence on dry surfaces [65]. |
| Enterococcus faecium | Polysaccharide Antigen, Pili | Adhesion forces modulated by surface glycopolymers; pili contribute to cell-cell cohesion [65]. | Surface polymers may shield adhesive molecules; pili are critical for aggregate formation in early biofilms [65]. |
The data reveal a clear trend: the presence of a thick, hydrated capsular polysaccharide generally correlates with a softer, more compliant cellular phenotype (lower Young's modulus), as prominently observed in K. pneumoniae [15]. This mechanical softness can enhance the contact area with surfaces, promoting adhesion. Furthermore, accessory structures like fimbriae and pili play a crucial role in modulating the capsule's organization and initiating direct, high-force contact with substrates [66] [15].
Diagram 1: AFM nanomechanics workflow for biofilm research.
Experimental Protocols for Key Investigations
Protocol: Quantifying Capsule Influence on Adhesion and Stiffness
This protocol is adapted from studies on K. pneumoniae [15].
- Objective: To determine the role of the polysaccharide capsule in bacterial nanomechanics by comparing wild-type (WT) and isogenic capsule-deficient mutant strains.
- Materials:
- WT and mutant bacterial strains (e.g., Δcps mutant).
- AFM with a liquid cell and cantilevers with spring constants of ~0.01-0.1 N/m.
- Freshly cleaved mica or glass substrates.
- Appropriate growth medium and phosphate-buffered saline (PBS).
- Procedure:
- Culture and Harvest: Grow bacterial strains to mid-logarithmic phase. Harvest cells by gentle centrifugation and wash twice in PBS to remove residual medium.
- Immobilization: Deposit a 50 µL droplet of the bacterial suspension (~10⁸ cells/mL) onto a poly-L-lysine-coated mica surface. Incubate for 30 minutes, then carefully rinse with PBS to remove non-adherent cells.
- AFM Imaging: Mount the sample in the AFM liquid cell filled with PBS. Using a soft cantilever, acquire topographic images in peak force quantitative nanomechanical mapping (PF-QNM) mode to visualize cell morphology and capsule presence.
- Force Curve Acquisition: On the top of at least 20 individual cells per strain, collect a grid of force-distance curves (e.g., 32x32 points over a 1x1 µm area).
- Data Analysis:
- Fit the retraction portion of the force curves to determine the maximum adhesion force (Fad).
- Fit the approaching (indentation) portion of the force curve with an appropriate contact mechanics model (e.g., Hertzian or Sneddon model) to extract the Young's modulus (E).
- Expected Outcome: The WT encapsulated strain will typically exhibit a lower average Young's modulus and altered adhesion profile compared to the stiffer, non-encapsulated mutant, demonstrating the capsule's role in mechanical compliance [15].
Protocol: Single-Cell Adhesion Force Spectroscopy
- Objective: To measure the specific adhesion forces between a pathogen and a relevant biological surface.
- Materials:
- Bacterial strain.
- AFM with tipless cantilevers.
- Colloidal probe or chemical linker (e.g., PEG-linker, polydopamine).
- Target substrate (e.g., epithelial cell line, coated surface).
- Procedure:
- Probe Functionalization: Attach a single live bacterial cell to the apex of a tipless cantilever using a bio-compatible adhesive like polydopamine or a flexible PEG-crosslinker.
- Substrate Preparation: Culture a monolayer of host cells or coat the substrate with a specific protein (e.g., fibronectin, collagen) on a Petri dish.
- Force Measurement: Approach the bacterium-functionalized probe to the substrate and record thousands of force-distance curves at multiple locations.
- Specificity Control: Repeat measurements in the presence of a soluble inhibitor (e.g., free sugar for lectin-mediated adhesion, antibody for a specific protein) to block specific interactions.
- Expected Outcome: The distribution of adhesion forces (seen as peaks in the retraction curve) reveals the strength and prevalence of bonds. A rightward shift in this distribution in the presence of an inhibitor confirms the specificity of the interaction [66].
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for AFM-Based Pathogen Nanomechanics
| Item | Function/Application | Technical Notes |
|---|---|---|
| Soft Cantilevers (k ≈ 0.01 - 0.1 N/m) | Force spectroscopy on live cells. | Pre-calibrate spring constants. Use silicon nitride for biocompatibility in liquid. |
| Functionalized Tips | Chemical Force Microscopy (CFM). | Tips coated with specific ligands (e.g., mannose, antibodies) to map receptor distribution. |
| PEG Crosslinkers | Covalently tether single bacterial cells to AFM cantilevers for SCFS. | Provides flexibility, reducing non-specific surface contact and allowing natural orientation. |
| Poly-L-Lysine | Electrostatic immobilization of cells onto mica/glass substrates. | A standard, simple method for creating a positively charged surface to trap negatively charged cells. |
| Polydopamine Adhesive | A versatile, strong bio-adhesive for attaching cells or proteins to AFM probes and substrates. | Can be used for both probe functionalization and surface coating; effective in aqueous environments. |
| DNA Origami Assay | Quantitative control substrate for assessing AFM tip effects on molecular interactions [67]. | Used to validate that imaging parameters do not artificially disrupt the system being studied. |
Diagram 2: Capsule and fimbriae roles in adhesion.
AFM-based nanomechanics provides an indispensable platform for quantitatively differentiating the biophysical properties of ESKAPE pathogens. The evidence clearly demonstrates that capsular polysaccharides are not merely passive protective barriers but are active mechanical elements that govern cellular stiffness, adhesion, and ultimately, biofilm initiation. The correlation between a soft, compliant capsule and enhanced biofilm formation in K. pneumoniae exemplifies a key finding enabled by this technology [15].
Future research directions will likely involve the deeper integration of AFM with complementary techniques like correlative fluorescence microscopy to link specific molecular events with nanomechanical responses in real-time [66]. The application of high-speed AFM will further allow researchers to visualize the dynamic rearrangement of surface structures during the adhesion process. As the field progresses, standardized protocols and control experiments, such as using DNA origami to quantify tip effects, will be crucial for generating robust and comparable data across laboratories [67]. Ultimately, a detailed understanding of pathogen nanomechanics will open new avenues for combating biofilm-related infections, potentially through the development of anti-adhesive therapies or materials engineered to resist colonization based on mechanical principles.
Bacterial biofilms represent a significant challenge in medical and industrial settings due to their inherent tolerance to antimicrobial agents. The development of non-biocidal strategies to prevent biofilm formation has emerged as a critical approach to combat biofilm-associated infections without promoting antibiotic resistance. This technical guide explores the groundbreaking research on bacterial capsular polysaccharides with antibiofilm activity, focusing on their distinctive electrokinetic and biophysical signatures. By integrating findings from atomic force microscopy (AFM) nanomechanics studies and systematic screening of polysaccharide libraries, we elucidate how specific physical properties—rather than particular chemical motifs—govern anti-adhesion capabilities. The characterization of a distinct electrokinetic signature associated with antibiofilm activity opens new perspectives for identifying or engineering non-biocidal surface-active macromolecules to control biofilm formation.
Bacterial biofilms are surface-attached communities that are difficult to eradicate due to their high tolerance to antimicrobial agents, making them a persistent problem on medical devices and industrial surfaces [16]. The prevention of biofilm-associated infections represents a major health and economic challenge, particularly in healthcare settings where indwelling medical devices are common sources of infection [15]. Traditional biocidal approaches using broad-spectrum antibiotics or heavy metals face limitations including the accumulation of dead bacteria and organic debris that reduces surface activity toward new incoming cells, and the concerning selection for antibiotic-resistant strains [16].
Non-antibiotic anti-adhesion strategies have emerged as promising alternatives that efficiently interfere with bacterial biofilm formation without promoting resistance [16]. Among these approaches, high molecular weight capsular polysaccharides released by various bacteria have demonstrated remarkable ability to prevent adhesion and subsequent biofilm formation by a wide range of Gram-positive and Gram-negative pathogens [16]. Unlike secreted bacterial antagonistic macromolecules such as colicins, toxins, or phages, these antibiofilm polysaccharides are non-biocidal and function by modifying surface properties including wettability, charge, and overall bacteria-surface contact dynamics [16].
The biophysical mechanisms through which polysaccharide capsules influence biofilm formation have been increasingly elucidated through nanomechanics approaches. AFM studies of pathogens such as Klebsiella pneumoniae have revealed that the organization of the capsule significantly influences bacterial adhesion and thereby biofilm formation [15]. Theoretical modeling of mechanical data alongside traditional biofilm assays has demonstrated that the structural organization of bacterial polysaccharide capsules plays a fundamental role in the initial stages of surface attachment [15] [24].
Despite these advances, the limited understanding of the chemical and structural bases of antibiofilm macromolecule activity has hindered their prophylactic application for bacterial biofilm control. This whitepaper synthesizes recent breakthroughs in identifying the electrokinetic signatures associated with antibiofilm activity, providing researchers with a framework for exploiting these biophysical properties in therapeutic and industrial applications.
Results and Data Analysis
Polysaccharide Screening and Anti-Biofilm Activity Identification
A comprehensive screening of 31 purified capsular polysaccharides of known composition and structure identified multiple compounds with non-biocidal activity against Escherichia coli and/or Staphylococcus aureus biofilms [16]. These polysaccharides were produced and purified from various strains of Streptococcus pneumoniae, Salmonella enterica serovar Typhi, Haemophilus influenzae, and Neisseria meningitidis, many of which are used as antigens in polysaccharide and glycoconjugate human vaccines [16].
Table 1: Antibiofilm Activity Profile of Selected Capsular Polysaccharides
| Polysaccharide | Activity Against E. coli | Activity Against S. aureus | Activity Spectrum |
|---|---|---|---|
| G2cps | Yes | Yes | Broad-spectrum |
| Vi | Yes | Yes | Broad-spectrum |
| MenA | Yes | Yes | Broad-spectrum |
| MenC | Yes | Yes | Broad-spectrum |
| PRP | Yes | Yes | Broad-spectrum |
| PnPS3 | Yes | Yes | Broad-spectrum |
| PnPS12F | Yes | No | Narrow-spectrum |
| PnPS18C | No | Yes | Narrow-spectrum |
| MenY | Yes | Yes | Broad-spectrum |
| MenW135 | Yes | Yes | Broad-spectrum |
The screening process employed static microtiter plate biofilm assays followed by crystal violet staining, with polysaccharides tested at equivalent concentrations (100 µg/mL) [16]. The results revealed distinct activity patterns, with some polysaccharides such as Vi, MenA, and MenC demonstrating broad-spectrum activity against both bacterial species, while others exhibited species-specific inhibition [16]. These findings were further validated using dynamic assays with continuous flow biofilm microfermentors, which confirmed the strong inhibitory effect of active polysaccharides like Vi on both E. coli and S. aureus biofilm formation [16].
Molecular Weight and Intrinsic Viscosity Correlations
The molecular weight (Mw) and intrinsic viscosity ([η]) of both active and inactive polysaccharides were determined using High Performance Size-Exclusion Chromatography coupled to Static Light Scattering (HPSEC-LS) [16]. Intrinsic viscosity reflects the contribution of a polysaccharide to the viscosity of the whole solution and depends on the conformation adopted by the polysaccharides in solution, which is mediated by various physicochemical parameters including electrostatic charges and their distribution within the macromolecular structure [16].
Table 2: Biophysical Properties of Active and Inactive Polysaccharides
| Polysaccharide | Molecular Weight (kDa) | Intrinsic Viscosity (dl/g) | Antibiofilm Activity |
|---|---|---|---|
| G2cps | 800 | >7 | Broad-spectrum |
| Vi | Not specified | >7 | Broad-spectrum |
| MenA | Not specified | >7 | Broad-spectrum |
| MenC | Not specified | >7 | Broad-spectrum |
| PnPS3 | Not specified | >7 | Broad-spectrum |
| PRP | Not specified | >7 | Broad-spectrum |
| PnPS12F | Not specified | Intermediate | Narrow-spectrum |
| PnPS18C | Not specified | Intermediate | Narrow-spectrum |
| Inactive Polysaccharides | Various | <7 | None |
This analysis revealed a remarkable correlation between intrinsic viscosity and broad-spectrum antibiofilm activity. All inactive macromolecules systematically displayed the lowest values of [η], whereas active polysaccharides were characterized by a high (>7 dl/g) intrinsic viscosity [16]. Polysaccharides with intermediate narrow-spectrum activity (PnPS18C and PnPS12F) covered a range of values between those measured for broad-spectrum active and non-active macromolecules [16].
The critical importance of molecular integrity was demonstrated through gradual reduction of G2cps polysaccharide size while preserving structural integrity using radical oxidation hydrolysis. Even minor reduction of polysaccharide size resulted in complete loss of G2cps antibiofilm activity, indicating that conservation of the polymer size is critical for antiadhesion properties [16].
Electrokinetic Properties of Antibiofilm Polysaccharides
The electrophoretic mobility of a subset of 21 capsular polysaccharides was measured and theoretically interpreted under applied electric field conditions [16]. This analysis revealed that active and inactive polysaccharide polymers display distinct electrokinetic properties, with all active macromolecules sharing high intrinsic viscosity features [16].
Despite the absence of specific molecular motifs associated with antibiofilm properties, the researchers identified that high density of electrostatic charges and permeability to fluid flow served as reliable criteria for predicting antibiofilm potential. Using these parameters, the research team successfully identified two additional capsular polysaccharides (MenY and MenW135) with broad-spectrum antibiofilm activity, confirming the predictive value of these biophysical characteristics [16].
Experimental Protocols
Polysaccharide Purification and Structural Characterization
Protocol 1: Polysaccharide Purification from Bacterial Cultures
Bacterial Strain Selection: Select appropriate bacterial strains known to produce the capsular polysaccharide of interest. Common sources include Streptococcus pneumoniae, Salmonella enterica serovar Typhi, Haemophilus influenzae, and Neisseria meningitidis [16].
Culture Conditions: Grow bacterial cultures under optimal conditions for polysaccharide production, typically in rich media with appropriate aeration and temperature control specific to each bacterial species.
Polysaccharide Extraction: Harvest bacterial cells during late exponential or early stationary phase by centrifugation. Extract capsular polysaccharides using one of the following methods:
- Heat Extraction: Incubate cell suspension at 50-65°C for 30-60 minutes followed by precipitation with ethanol or cetrimonium bromide [16].
- Enzymatic Digestion: Treat cell pellets with proteases to remove protein contaminants followed by dialysis and precipitation.
- Chemical Extraction: Use organic solvents or detergents to solubilize polysaccharides while maintaining structural integrity.
Purification Steps:
- Perform size-exclusion chromatography using Sepharose or Sephacryl matrices.
- Utilize ion-exchange chromatography for charged polysaccharides.
- Remove contaminants and lipopolysaccharides through multiple precipitation steps and ultracentrifugation.
- Confirm purity using SDS-PAGE and protein quantification assays.
Quality Control: Verify polysaccharide integrity through molecular weight analysis using HPSEC-LS and monosaccharide composition analysis by HPAEC-PAD [16].
Protocol 2: Structural Characterization of Polysaccharides
Composition Analysis:
- Perform monosaccharide analysis using High-Performance Anion-Exchange Chromatography with Pulsed Amperometric Detection (HPAEC-PAD) [16].
- Conduct methylation analysis to determine glycosidic linkages.
- Identify non-carbohydrate components (e.g., O-acetyl, glycerol phosphate, pyruvate) using colorimetric assays and chromatography.
Nuclear Magnetic Resonance (NMR) Spectroscopy:
- Acquire 1H, 13C, and 31P NMR spectra on purified polysaccharide samples dissolved in D2O [16].
- Use two-dimensional NMR techniques (COSY, TOCSY, NOESY, HSQC, HMBC) for complete structural assignment.
- Determine repeating unit structure through analysis of spin systems and inter-residue correlations.
Molecular Weight Determination:
- Perform High Performance Size-Exclusion Chromatography coupled to Static Light Scattering (HPSEC-LS) to determine weight-average molecular weight (Mw) and polydispersity [16].
- Calculate intrinsic viscosity ([η]) from SEC-LS data using the relationship [η] = (η−ηο)/(ηο ϕ), where ηο is the solution viscosity without polysaccharide and ϕ is the volume fraction of polysaccharides [16].
Biofilm Assays and Assessment of Antibiofilm Activity
Protocol 3: Static Microtiter Plate Biofilm Assay
Bacterial Strain Preparation:
- Grow test organisms (E. coli, S. aureus, or other relevant pathogens) overnight in appropriate media.
- Dilute fresh cultures to approximately 1×10^6 CFU/mL in fresh medium supplemented with or without polysaccharides at test concentration (typically 100 µg/mL) [16].
Biofilm Formation:
- Dispense 200 µL of bacterial suspension into 96-well polystyrene microtiter plates.
- Include controls: medium alone (negative control), bacteria without polysaccharides (positive control), and polysaccharides alone (background control).
- Incubate plates under optimal growth conditions for 24-48 hours without agitation.
Biofilm Quantification:
- Carefully remove planktonic cells by inverting and tapping the plate.
- Wash adherent cells gently with phosphate-buffered saline (PBS) to remove loosely attached cells.
- Fix biofilms with 99% methanol for 15 minutes, then air dry.
- Stain with 0.1% crystal violet solution for 15-20 minutes.
- Wash extensively with distilled water to remove excess stain.
- Destain with 33% glacial acetic acid or ethanol-acetone mixture (80:20).
- Transfer 125 µL of destained solution to a new microtiter plate.
- Measure absorbance at 570-600 nm using a microplate reader.
Data Analysis:
- Calculate percentage inhibition relative to positive control: % Inhibition = [1 - (ODsample - ODnegative control)/(ODpositive control - ODnegative control)] × 100.
- Perform statistical analysis using appropriate tests (e.g., Student's t-test, ANOVA with post-hoc tests).
Protocol 4: Dynamic Biofilm Assay Using Continuous Flow Microfermentors
System Setup:
- Assemble continuous flow biofilm systems with appropriate growth chambers (e.g., flow cells, tube reactors).
- Sterilize the system by autoclaving or chemical sterilization.
- Connect to medium reservoir with peristaltic pump for controlled flow.
Biofilm Formation Under Flow Conditions:
- Inoculate the system with bacterial suspension (1×10^6 CFU/mL) and allow initial attachment without flow for 1-2 hours.
- Initiate continuous flow of fresh medium supplemented with test polysaccharides at desired concentration.
- Maintain constant flow rate (typically 0.2-0.5 mL/min) to create laminar flow conditions.
- Incubate for 24-72 hours at appropriate temperature.
Biofilm Harvesting and Analysis:
- Carefully remove biofilm samples from the chamber surfaces at predetermined time points.
- Quantify biofilm biomass through:
- Crystal violet staining as described in Protocol 3
- Viable cell counts by sonicating biofilm and plating serial dilutions
- Direct microscopic examination
- Perform confocal laser scanning microscopy for structural analysis if needed.
Protocol 5: Biocompatibility and Non-biocidal Activity Assessment
Bacterial Viability Assay:
- Expose bacterial cells to polysaccharides at working concentrations (100 µg/mL) in liquid culture [16].
- Monitor growth curves by measuring optical density at 600 nm over 24 hours.
- Compare growth kinetics between treated and untreated cultures.
Colony Forming Unit (CFU) Enumeration:
- Plate serial dilutions of polysaccharide-treated and untreated bacterial cultures on solid media.
- Incubate plates overnight and count resulting colonies.
- Calculate CFU/mL and compare between conditions.
Membrane Integrity Assessment:
- Perform Live/Dead staining using SYTO9 and propidium iodide.
- Analyze by fluorescence microscopy or flow cytometry.
- Confirm non-biocidal activity by demonstrating high percentage of viable cells in treated cultures comparable to untreated controls [16].
AFM Nanomechanics Measurements
Protocol 6: In Situ Nanomechanical Measurements of Bacterial Cells
Sample Preparation:
- Grow bacterial cells (wild type and specific mutants) to mid-exponential phase.
- Gently harvest cells by centrifugation (2000-5000 × g for 5 minutes) to preserve surface structures.
- Wash cells in appropriate buffer (e.g., PBS or growth medium).
- Immobilize cells on freshly cleaned substrate surfaces (e.g., poly-L-lysine coated glass coverslips, gelatin-coated surfaces) by allowing adhesion for 15-30 minutes.
- Carefully rinse with buffer to remove non-adherent cells.
- Keep samples hydrated throughout preparation and measurement.
AFM Instrument Setup:
- Mount appropriate cantilevers with known spring constants (typically 0.01-0.1 N/m for bacterial measurements).
- Calibrate cantilever sensitivity using clean, rigid surface (e.g., glass or mica).
- Determine exact spring constant using thermal noise method or reference cantilever method.
- Select probes with colloidal tips or sharp tips depending on measurement type (nanomechanics vs. high-resolution imaging).
Force Spectroscopy Measurements:
- Approach immobilized cells in liquid environment at controlled temperature.
- Acquire force-distance curves at multiple locations on individual cells (minimum 100 curves per cell, 10+ cells per condition).
- Set appropriate trigger points (typically 0.5-1 nN) to avoid cell damage.
- Maintain consistent approach and retraction speeds (0.5-1 μm/s).
- Include measurements on bare substrate as control.
Data Analysis:
- Process force curves to extract mechanical parameters:
- Adhesion Force: Minimum value in retraction curve.
- Elastic Modulus: Fit approach curve with appropriate contact mechanics model (Hertz, Sneddon, or JKR model).
- Turgor Pressure: Analyze using theoretical models of bacterial cell mechanics.
- Capsule Thickness: Determine from contact point in force curves.
- Perform statistical analysis to compare parameters between different bacterial strains or conditions.
- Correlate nanomechanical data with biofilm formation capabilities from parallel assays [15] [24].
- Process force curves to extract mechanical parameters:
Visualization of Experimental Workflows and Relationships
Antibiofilm Polysaccharide Screening Workflow
Figure 1: Comprehensive workflow for screening and characterizing antibiofilm polysaccharides, integrating purification, structural analysis, and functional assessment.
Biophysical Properties and Anti-Biofilm Activity Relationship
Figure 2: Relationship between biophysical properties of polysaccharides and their anti-biofilm mechanisms, highlighting the connection between specific characteristics and functional outcomes.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Research Reagents and Materials for Anti-Biofilm Polysaccharide Studies
| Category | Specific Items | Function/Application | Technical Notes |
|---|---|---|---|
| Bacterial Strains | Escherichia coli (UPEC strains), Staphylococcus aureus, Klebsiella pneumoniae (wild type and mutants) | Biofilm formation assays, adhesion studies, polysaccharide production | Select clinically relevant isolates with well-characterized biofilm phenotypes; include isogenic mutants for mechanistic studies [15] |
| Polysaccharide Sources | Purified capsular polysaccharides from S. pneumoniae, S. Typhi (Vi), H. influenzae (PRP), N. meningitidis (MenA, MenC, MenY, MenW135) | Anti-biofilm activity screening, structure-function studies | Source from commercial suppliers or purify in-house; verify structure and purity through compositional analysis and NMR [16] |
| Analytical Instruments | HPSEC-LS (High Performance Size-Exclusion Chromatography with Light Scattering), HPAEC-PAD (High-Performance Anion-Exchange Chromatography), NMR Spectrometer | Molecular weight determination, monosaccharide composition analysis, structural characterization | HPSEC-LS provides both Mw and intrinsic viscosity ([η]); HPAEC-PAD enables sensitive detection of neutral and acidic sugars without derivatization [16] |
| Biofilm Assessment Tools | 96-well microtiter plates, crystal violet, continuous flow microfermentors, confocal laser scanning microscope | Biofilm quantification, dynamic biofilm analysis, 3D structural characterization | Combine static and dynamic assays for comprehensive assessment; use COMSTAT or similar software for image analysis of biofilm architecture |
| Nanomechanics Equipment | Atomic Force Microscope with liquid cell, colloidal probes, temperature controller | In situ nanomechanical measurements of bacterial cells, adhesion force quantification | Use appropriate cantilever spring constants (0.01-0.1 N/m); apply suitable contact mechanics models for data analysis; maintain physiological conditions [15] [24] |
| Electrokinetic Analysis | Zeta potential analyzer, electrophoretic mobility measurement system | Characterization of surface charge properties, evaluation of electrokinetic signatures | Measure under physiological ionic strength conditions; correlate with antibiofilm activity [16] |
Discussion and Future Perspectives
The identification of electrokinetic signatures and specific biophysical properties associated with antibiofilm activity represents a paradigm shift in the development of non-biocidal anti-biofilm strategies. The finding that high intrinsic viscosity (>7 dl/g), high electrostatic charge density, and permeability to fluid flow serve as reliable predictors of broad-spectrum antibiofilm activity provides researchers with valuable criteria for screening and designing novel anti-adhesion polymers [16]. This biophysical approach complements traditional structure-activity relationship studies that focus primarily on chemical motifs.
The integration of AFM nanomechanics into biofilm research has been particularly transformative, enabling in situ measurements of bacterial cell surfaces and their mechanical responses to environmental conditions [15] [24]. These studies have revealed that bacterial capsules behave as responsive polymer hydrogels that adapt to osmotic stress, functioning as "ion sponges" that dampen the impact of environmental challenges [24]. This fundamental understanding of capsule biomechanics provides insight into how capsular polysaccharides might function in anti-biofilm applications by modifying the interface between bacterial cells and surfaces.
The demonstrated importance of molecular size integrity for antibiofilm activity [16] highlights the necessity of careful handling during polysaccharide purification and storage to prevent degradation. The loss of activity with even minor size reduction suggests that these polymers function through a mechanism requiring critical spatial dimensions, possibly related to their ability to form protective hydration layers or interfere with adhesion receptors through steric hindrance effects.
Future research directions should focus on several key areas:
- Engineering of Synthetic Analogues: Developing synthetic polymers with optimized biophysical properties that mimic the electrokinetic signatures of natural antibiofilm polysaccharides while offering improved stability and production scalability.
- Surface Coating Technologies: Incorporating active polysaccharides or their synthetic mimics into surface coatings for medical devices to prevent biofilm formation without eliciting resistance.
- Combination Therapies: Exploring synergistic effects between antibiofilm polysaccharides and conventional antimicrobials to enhance efficacy while reducing antibiotic concentrations.
- In Vivo Validation: Extending promising in vitro findings to appropriate animal models to assess therapeutic potential and biocompatibility.
The characterization of distinct electrokinetic signatures associated with antibiofilm activity opens new avenues for identifying or engineering non-biocidal surface-active macromolecules. This approach has significant potential for controlling biofilm formation in medical and industrial settings while circumventing the selective pressure for antibiotic resistance associated with traditional biocidal strategies.
Cross-Validation with Other Anti-Biofilm Strategies (Nanoparticles, Enzymes)
Bacterial biofilms are structured communities of microbial cells enclosed in a self-produced extracellular polymeric matrix that demonstrate remarkable tolerance to antimicrobial agents, contributing significantly to persistent infections and treatment failures [8] [68]. The global health impact of biofilm-associated antimicrobial resistance is profound, with an estimated 7 million deaths annually linked to antimicrobial resistance, a figure projected to rise to 10 million by 2050 without effective interventions [8]. Within this challenging landscape, capsular polysaccharides (CPS) have emerged as critical players in both biofilm formation and inhibition, making them a focal point for therapeutic investigation [16] [69].
The integration of Atomic Force Microscopy (AFM) nanomechanics into biofilm research provides unprecedented quantitative analysis of the biomechanical properties of biofilms and their constituent components at the nanoscale [61]. However, to validate findings and develop comprehensive therapeutic strategies, researchers must contextualize AFM data within the broader arsenal of anti-biofilm approaches. This technical guide examines how nanoparticle-based and enzymatic anti-biofilm strategies complement AFM nanomechanics research on capsular polysaccharides, providing methodologies for cross-validation and establishing mechanistic correlations between nanomechanical properties and anti-biofilm efficacy.
Atomic Force Microscopy in Biofilm Nanomechanics
Fundamental AFM Methodologies for Biofilm Characterization
Atomic Force Microscopy offers diverse operational modes for interrogating biofilm systems. In contact mode, the tip maintains continuous contact with the surface, while tapping mode (intermittent contact) reduces lateral forces and is preferred for soft biological samples [61]. Phase imaging, captured simultaneously with topographical data, provides qualitative distinction between materials on heterogeneous surfaces based on mechanical properties [61].
Key AFM Methodologies:
- Force-Distance Curve Analysis: Measures interaction forces between the AFM tip and sample surface, revealing adhesive properties, binding forces, and molecular recognition events [61].
- Nanoindentation: Quantifies mechanical properties including elastic modulus and turgor pressure by comparing force curves on reference hard surfaces and biofilm samples, typically using Hertz model analysis [61].
- Single-Cell Immobilization: Essential for reliable imaging and force measurement, achieved through mechanical entrapment in porous media or chemical fixation using poly-L-lysine or other benign adhesives [61].
AFM Applications in Capsular Polysaccharide Research
AFM enables direct correlation between CPS structural features and their nanomechanical properties. High-resolution imaging reveals CPS architecture and distribution, while force spectroscopy quantifies their contribution to cell-surface adhesion and intercellular cohesion within biofilms [61]. These measurements provide foundational data for cross-validation with other anti-biofilm strategies by establishing baseline mechanical properties that can be monitored during therapeutic interventions.
Nanoparticle-Based Anti-Biofilm Strategies
Mechanisms of Nanoparticle-Mediated Biofilm Disruption
Nanoparticles combat biofilms through multiple mechanisms, offering distinct advantages for cross-validation with AFM nanomechanical studies. Their high surface-area-to-volume ratio and tunable surface chemistry enable enhanced penetration into biofilm matrices and targeted interactions with CPS components [8] [68].
Table 1: Nanoparticle Types and Their Anti-Biofilm Mechanisms
| Nanoparticle Type | Primary Anti-Biofilm Mechanism | Key Properties | Relevance to CPS Research |
|---|---|---|---|
| Silver (Ag) NPs | Membrane disruption, ROS generation, QS inhibition [68] | Size: 20-40 nm, Concentration: 10-100 μg/mL [68] | Alters CPS mechanical properties; reduces matrix cohesion |
| Functionalized Polymer NPs | Drug delivery, matrix penetration, synergistic combination [8] | Biodegradable, surface functionalization | Targeted CPS disruption; enables controlled release of CPS-active compounds |
| Metal Oxide NPs (ZnO, CuO) | ROS generation, mechanical stress [68] | Photocatalytic activity, ion release | Indirect CPS modification through oxidative damage |
| Mechano-bactericidal Nanostructures | Physical piercing of cell membranes [70] | Nanostructured surfaces with ~200 nm pillars | Physical bypass of CPS protection; induces mechanical failure |
Experimental Protocols for Nanoparticle Biofilm Studies
Protocol: Evaluating Nanoparticle Anti-Biofilm Efficacy
- Biofilm Cultivation: Grow biofilms in flow cells or microtiter plates for 24-72 hours using appropriate media [8].
- NP Treatment: Apply nanoparticle suspensions (1-100 μg/mL concentration range) for 4-24 hours [68].
- Viability Assessment: Quantify metabolic activity using resazurin assay or colony-forming unit (CFU) counts [71].
- Biomass Quantification: Measure total biofilm biomass using crystal violet staining [16].
- Structural Analysis: Employ confocal laser scanning microscopy (CLSM) with fluorescent markers to assess biofilm architecture [8].
Protocol: AFM-NP Cross-Validation
- Pre-Treatment Characterization: Perform AFM nanoindentation and adhesion force mapping on untreated biofilms to establish baseline mechanical properties [61] [72].
- Controlled NP Application: Apply sub-inhibitory concentrations of nanoparticles to monitor progressive changes.
- Post-Treatment AFM Analysis: Quantify alterations in elastic modulus, adhesion forces, and surface topography [61].
- Correlation Analysis: Establish statistical relationships between NP-induced mechanical changes and anti-biofilm efficacy.
Figure 1: Nanoparticle Anti-Biofilm Mechanisms and AFM Cross-Validation Pathways
Enzymatic Anti-Biofilm Approaches
Enzyme Classes and Their Targets in Biofilm Matrices
Enzymatic strategies target specific components of the biofilm extracellular polymeric substance (EPS), including CPS, proteins, and extracellular DNA, offering precise mechanistic insights complementary to AFM nanomechanics [73].
Table 2: Enzymes for Biofilm Matrix Degradation
| Enzyme Class | Specific Examples | Target Substrate | Mechanical Outcome |
|---|---|---|---|
| Glycoside Hydrolases | Dispersin B, Levan hydrolase, Cellulase [73] | Poly-N-acetylglucosamine (PNAG), levan, cellulose [73] | Reduced cohesion, decreased stiffness |
| Proteases | Lysostaphin, various proteases [73] | Matrix proteins, surface adhesins | Weakened structural integrity, reduced adhesion |
| Deoxyribonucleases | DNase I [73] | Extracellular DNA (eDNA) | Loss of structural stability, enhanced detachment |
| Polysaccharide Lyases | Alginate lyase [73] | Alginate and other uronic acid-containing polymers | Reduced viscosity, matrix dissolution |
Experimental Protocols for Enzymatic Biofilm Disruption
Protocol: Enzyme Susceptibility Testing
- Biofilm Growth: Cultivate biofilms under conditions that promote robust matrix production (48-72 hours) [73].
- Enzyme Preparation: Prepare enzyme solutions in appropriate buffers at concentrations of 0.1-100 μg/mL, with controls using heat-inactivated enzymes [73].
- Treatment Application: Apply enzymes to pre-formed biofilms for 1-6 hours at optimal temperature and pH.
- Efficacy Assessment: Quantify biofilm removal by crystal violet staining, CFU enumeration, or microscopy [16] [73].
- Matrix Component Analysis: Monitor release of specific monosaccharides (for polysaccharidases) or nucleotides (for DNases) to confirm target engagement [73].
Protocol: AFM-Enzyme Cross-Validation
- Real-Time Mechanical Monitoring: Employ AFM to track changes in biofilm viscoelastic properties during enzyme treatment using time-series nanoindentation [61] [72].
- Adhesion Mapping: Quantify reductions in adhesion forces following enzyme exposure, particularly for adhesin-targeting proteases.
- Correlative Microscopy: Combine AFM with fluorescence microscopy using enzyme substrates tagged with fluorescent probes.
- Single-Molecule Force Spectroscopy: Measure direct enzyme-polysaccharide interactions at the molecular level [61].
Integrated Cross-Validation Framework
Correlating Nanomechanical Properties with Anti-Biofilm Efficacy
Successful cross-validation requires establishing quantitative relationships between AFM-derived mechanical parameters and conventional anti-biofilm efficacy metrics.
Table 3: Cross-Validation Parameters for Anti-Biofilm Strategies
| AFM Mechanical Parameter | Experimental Method | Correlation with Anti-Biofilm Efficacy | Interpretation Guidelines |
|---|---|---|---|
| Elastic (Young's) Modulus | Nanoindentation using Hertz model [61] [72] | Inverse correlation with biofilm detachment | >50% reduction indicates significant matrix disruption |
| Adhesion Force | Single-cell force spectroscopy, force-volume mapping [61] | Direct correlation with initial attachment strength | >60% reduction suggests impaired surface colonization |
| Viscoelastic Parameters (creep compliance, relaxation time) | Stress relaxation tests [72] | Correlates with biofilm removal under shear | Increased compliance enhances susceptibility to fluid shear |
| Surface Roughness | Topographical imaging [61] | Changes indicate structural reorganization | Increased roughness often precedes biofilm detachment |
Comprehensive Experimental Workflow for Cross-Validation
Figure 2: Integrated Cross-Validation Workflow for Anti-Biofilm Strategies
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 4: Key Research Reagents for Anti-Biofilm Studies
| Reagent/Material | Function | Application Notes | References |
|---|---|---|---|
| Polydimethylsiloxane (PDMS) Stamps | Cell immobilization for AFM | Enables reproducible single-cell analysis without chemical modification | [61] |
| Crystal Violet Solution | Biofilm biomass quantification | 0.1% solution for microtiter plate assays; limited to total biomass | [16] |
| Resazurin Sodium Salt | Metabolic activity assessment | 0.15-0.5 mg/mL in buffer; measures viability without biofilm disruption | [71] |
| Recombinant Dispersin B | PNAG-specific glycosidase | 1-100 μg/mL for matrix degradation; specific for β-1,6-linked GlcNAc | [73] |
| Functionalized Nanoparticles | Targeted biofilm penetration | Ag, ZnO, or polymer NPs with surface modifiers for enhanced diffusion | [68] |
| Hertz Model Analysis Software | Nanomechanical property calculation | Requires input of tip geometry, Poisson's ratio (typically 0.5 for biofilms) | [61] |
| Extracellular Polysaccharide Standards | Matrix component reference | Commercial CPS preparations for method validation and controls | [16] [69] |
The cross-validation of AFM nanomechanics with nanoparticle and enzymatic anti-biofilm strategies represents a powerful paradigm for advancing biofilm research. By establishing quantitative relationships between nanomechanical properties and therapeutic efficacy, researchers can bridge the gap between fundamental science and clinical applications. The integrated framework presented in this guide enables comprehensive characterization of anti-biofilm mechanisms, from molecular-level interactions to macroscopic outcomes. As the field progresses, standardization of mechanical testing protocols and shared databases of mechanical properties will be essential for comparing results across studies and accelerating the development of novel anti-biofilm therapeutics targeting capsular polysaccharides and other matrix components [72].
Translating Nanomechanical Insights into Anti-Adhesion Therapeutic Strategies
The formation of bacterial biofilms on biotic and abiotic surfaces represents a significant challenge in healthcare, particularly on indwelling medical devices where biofilms can lead to persistent hospital-acquired infections [15]. At the heart of this process lies the intricate role of bacterial capsular polysaccharides—gel-like polymeric structures that envelop bacterial cells and mediate their initial attachment to surfaces. While the biochemical properties of these capsules have been studied for decades, recent advances in atomic force microscopy (AFM) nanomechanics have revealed the crucial mechanical aspects of biofilm development that were previously inaccessible to researchers [15] [24]. This technical guide explores how nanomechanical insights, particularly from AFM studies of pathogens like Klebsiella pneumoniae, are illuminating novel therapeutic pathways to disrupt the initial adhesion stage of biofilm formation—a critical window of opportunity for intervention [15].
The polysaccharide capsule functions not merely as a static protective barrier but as a dynamic responsive polymer hydrogel that mechanically adapts to environmental conditions and surface properties [24]. Through in situ AFM measurements, researchers have demonstrated that this capsule acts as an "ion sponge" to dampen the impact of osmotic stress, highlighting its mechanoresponsive behavior [24]. This mechanical adaptability directly influences bacterial adhesion capacity and subsequent biofilm formation, providing new targets for anti-adhesion strategies that operate at the nanoscale.
Nanomechanical Mechanisms of Bacterial Adhesion
Structural Organization of Capsular Polysaccharides
The structural organization of the capsular polysaccharide layer, rather than merely its presence or biochemical composition, has been identified as a critical factor governing bacterial adhesion. AFM nanomechanics studies comparing wild-type and mutant Klebsiella pneumoniae strains have revealed that capsule organization significantly influences the initial stages of surface attachment [15]. Theoretical modeling of mechanical data from these studies demonstrates that the spatial arrangement and surface presentation of polysaccharide chains determine adhesion probability and strength [15].
Notably, the organization of the capsule is mechanically regulated by the presence of type 3 fimbriae, which appear to structurally arrange the polysaccharide layer to optimize its adhesive properties [15]. This interplay between proteinaceous fimbriae and polysaccharide capsules creates a sophisticated adhesion system that responds to surface properties and environmental conditions. Mutant strains lacking properly organized capsules demonstrate markedly reduced biofilm formation capabilities despite possessing the biochemical components of polysaccharide production, underscoring the mechanical—not just chemical—importance of structural organization [15].
Force-Dependent Adhesion Mechanisms
AFM-based force spectroscopy has uncovered several fundamental mechanisms through which capsular polysaccharides mediate adhesion:
Polymer brush dynamics: The capsular polysaccharides function as a polymer brush layer that exhibits distance-dependent interactions with surfaces. At sufficient separation distances, the polymer chains exert entropic repulsion forces, while at closer ranges, attractive forces dominate through van der Waals interactions and hydrogen bonding [24] [74].
Energy dissipation: During adhesion events, the capsule behaves as a viscoelastic hydrogel that dissipates mechanical energy through reversible deformation and polymer chain rearrangement. This energy dissipation capability enhances adhesion stability by reducing the mechanical forces that would otherwise detach the bacterium from the surface [24].
Adhesive unfolding: Certain polysaccharide structures undergo force-induced conformational changes that expose previously hidden adhesive domains. This mechanical unfolding response effectively strengthens adhesion under shear stress conditions, facilitating firm attachment in dynamic environments like the bloodstream or urinary tract [75].
Table 1: Key Nanomechanical Parameters of Bacterial Adhesion Measured by AFM
| Parameter | Description | Typical Range | Therapeutic Significance |
|---|---|---|---|
| Adhesion Force | Maximum force required to detach bacterium from surface | 0.1-5 nN [52] | Determines efficacy of mechanical disruption strategies |
| Adhesion Energy | Total work required for complete detachment | 10-500 aJ [52] | Predicts stability of attachment under flow conditions |
| Rupture Length | Distance over which adhesive bonds break | 10-500 nm [52] | Indicates polymer extensibility and bond complexity |
| Elastic Modulus | Stiffness of bacterial cell surface | 0.1-5 GPa [24] | Influences contact area and adhesion probability |
Advanced AFM Methodologies for Nanomechanical Analysis
Bimodal AFM with Nonlinear Response Analysis
Traditional AFM techniques measure cantilever response at a single frequency, providing limited information about material properties. Bimodal AFM significantly enhances material contrast by exciting and measuring two cantilever eigenmodes simultaneously [76]. This approach has been further refined through analysis of nonlinear response to the bimodal drive, measuring amplitude and phase at harmonics and mixing frequencies [76].
The technical implementation involves exciting the cantilever at two frequencies (f₁ and f₂) near its first two flexural resonances—for example, 78.5 kHz and 500.5 kHz [76]. The nonlinear tip-surface interaction generates intermodulation products at frequencies mathematically described as f = nf₁ + mf₂ (where n and m are integers) [76]. By measuring the response at up to 17 frequencies simultaneously, researchers can achieve an almost threefold improvement in material discrimination capability compared to standard bimodal operation [76].
This enhanced discrimination is particularly valuable for studying the mechanical heterogeneity of bacterial capsules, which may contain localized domains with distinct adhesive properties that are obscured in single-frequency measurements. The approach allows for quantitative material property mapping without increasing the applied force to the surface, thereby preserving the native structure of delicate biological samples [76].
Force Spectroscopy and Single-Molecule AFM
Force spectroscopy enables the quantitative measurement of specific interactions at the single-molecule level [75] [74]. In this methodology, the AFM tip is functionalized with specific ligands, surface proteins, or polysaccharides, and force-distance curves are recorded as the tip approaches and retracts from the bacterial surface [75] [74]. The resulting data reveal the unbinding forces of specific molecular interactions, kinetic parameters, and the mechanical properties of individual polysaccharide chains [75].
For capsule-specific measurements, several technical approaches have been developed:
Single polysaccharide detection: AFM tips can be used to mechanically manipulate individual polysaccharide molecules on live bacteria, providing information about their conformation, elasticity, and attachment points [24]. This has revealed that certain capsular polysaccharides can undergo substantial stretching before detachment, exhibiting remarkable mechanical resilience.
Adhesion force mapping: By recording force curves at multiple points across the bacterial surface, researchers can create spatial maps of adhesion forces with nanometer resolution [75]. This technique has revealed that bacterial adhesion is not uniform but concentrated in specific nanodomains that may represent privileged sites for initial surface attachment.
Single-mforce spectroscopy: This technique measures the forces required to stretch single polymer molecules or to break single bonds, providing fundamental insights into the mechanical strength of adhesive bonds [74].
Diagram 1: AFM Nanomechanics Workflow for Anti-Adhesion Therapeutic Development
Experimental Protocols for AFM-Based Adhesion Studies
Bacterial Immobilization and Sample Preparation
Proper sample preparation is critical for obtaining physiologically relevant nanomechanical data. The following protocol ensures appropriate bacterial immobilization while preserving capsular integrity:
Substrate selection: Use freshly cleaved mica, highly ordered pyrolytic graphite (HOPG), or functionalized glass coverslips as substrates [75]. Mica provides an atomically flat, negatively charged surface suitable for most bacterial strains.
Surface functionalization: For specific adhesion studies, functionalize substrates with target surfaces (e.g., collagen, fibronectin, polymer coatings) to mimic clinical relevant materials [52].
Bacterial immobilization: Apply 10-50 μL of bacterial suspension (OD₆₀₀ ≈ 0.1-0.5) to the substrate and allow to adhere for 10-30 minutes. For weaker adhering strains, use poly-L-lysine coating or gentle centrifugation (500 × g for 2 minutes) to enhance immobilization [75].
Washing: Gently rinse with appropriate buffer (e.g., PBS or growth medium) to remove non-adherent cells while preserving capsule integrity. Avoid excessive shear forces that might damage the polysaccharide layer.
Environmental control: Perform AFM measurements in liquid environment using appropriate fluid cells. Maintain temperature at 37°C for physiologically relevant conditions when possible [75].
AFM Force Curve Acquisition and Analysis
The acquisition and interpretation of force curves follows a standardized methodology:
Cantilever selection: Use soft cantilevers with spring constants of 0.01-0.1 N/m for adhesion measurements to ensure sufficient sensitivity while maintaining stability [74]. Colloidal probes with spherical tips may be used for more quantitative surface force measurements [74].
Approach-retract cycling: Program the piezoelectric scanner to approach and retract from the surface at constant velocity (typically 0.5-1 μm/s). Collect data at a sampling rate sufficient to capture molecular rupture events (≥2 kHz) [74].
Adhesion force calculation: Convert photodiode voltage to force using the cantilever spring constant (from thermal tuning or other calibration method) and the optical lever sensitivity [74]. The adhesion force is determined from the maximum negative force during retraction.
Rupture event analysis: Identify discrete rupture events in the retraction curve as sudden decreases in force. Calculate rupture lengths from the distance between events and adhesion energy from the area under the force-distance curve during retraction [52] [74].
Statistical analysis: Collect at least 100-200 force curves from multiple cells and locations to ensure statistical significance. Present data as mean ± standard deviation or as probability density distributions [52].
Table 2: Research Reagent Solutions for AFM Biofilm Studies
| Reagent/Category | Specific Examples | Function/Application | Technical Considerations |
|---|---|---|---|
| Substrates | Freshly cleaved mica, HOPG, functionalized glass [75] | Provides atomically flat surface for immobilization | Mica: negatively charged; HOPG: hydrophobic; Glass: versatile for functionalization |
| Functionalization | Poly-L-lysine, collagen, fibronectin, polymer coatings [52] | Enhances bacterial immobilization or mimics clinical surfaces | Concentration and incubation time critical for reproducibility |
| Cantilevers | Soft silicon nitride cantilevers (k=0.01-0.5 N/m), colloidal probes [74] | Measures forces with appropriate sensitivity | Spring constant calibration essential for quantitative measurements |
| Buffers | PBS, growth medium, Tris-HCl [75] | Maintains physiological conditions during measurement | Ionic strength affects electrostatic interactions and adhesion forces |
| Bacterial Strains | Wild-type vs. capsule-deficient mutants [15] | Identifies capsule-specific contributions to adhesion | Isogenic mutants provide most reliable comparisons |
Translating Nanomechanical Data into Therapeutic Strategies
Targeting Capsular Organization and Mechanics
The insights gained from AFM nanomechanics studies point to several promising therapeutic approaches for disrupting biofilm formation:
Capsule-disorganizing agents: Since the organization of the capsule—not merely its presence—governs adhesion, therapeutic strategies could target the proteins (like those in type 3 fimbriae) that structurally arrange the polysaccharide layer [15]. Small molecules that disrupt the capsule-fimbriae interaction could reduce adhesion without the evolutionary pressure associated with bactericidal agents.
Mechanically-responsive materials: The discovery that the capsule behaves as a responsive polymer hydrogel suggests that surfaces with specific nanomechanical properties could be designed to resist bacterial attachment [24]. Materials with tailored stiffness, viscoelasticity, or topography could present surfaces that are mechanically incompatible with capsule adhesion mechanisms.
Polymer-based anti-adhesives: Non-toxic polymer solutions could be developed to occupy the capsule's adhesive domains or alter its mechanical properties. These "molecular decoys" would prevent bacterial attachment to clinical surfaces without directly threatening bacterial survival, potentially reducing the development of resistance [24].
Quantitative Framework for Anti-Adhesion Therapeutics
The nanomechanical parameters obtained through AFM provide a quantitative framework for developing and evaluating anti-adhesion strategies:
Diagram 2: From Nanomechanical Insights to Therapeutic Strategies Translation Pathway
Adhesion force reduction: Effective anti-adhesion therapeutics should demonstrate a statistically significant reduction in measured adhesion force (target: >50% reduction) in AFM measurements [52].
Adhesion energy decrease: Successful strategies should lower the total work of adhesion (adhesion energy) by disrupting the multiple bond formations that occur during capsule-mediated attachment [52].
Altered mechanical properties: Therapeutic approaches that increase capsule stiffness or reduce its viscoelastic dissipation capacity may reduce adhesion efficiency by limiting contact area and energy absorption during attachment [24].
The translation of nanomechanical insights into anti-adhesion therapeutic strategies represents a paradigm shift in biofilm prevention. By targeting the mechanical rather than purely biochemical aspects of bacterial adhesion, this approach offers promising avenues for combating device-related infections while potentially reducing selective pressure for antibiotic resistance. The methodologies outlined in this technical guide—particularly advanced AFM techniques like bimodal operation with nonlinear analysis and single-molecule force spectroscopy—provide researchers with powerful tools to quantify adhesion parameters and validate therapeutic approaches at the nanoscale.
As the field progresses, the integration of nanomechanical data with computational modeling and materials science will further accelerate the development of targeted anti-adhesion therapeutics. The quantitative framework established through AFM studies of capsular polysaccharides provides a solid foundation for this multidisciplinary effort, pointing toward a future where mechanical intervention becomes a standard approach in infectious disease management.
Conclusion
The integration of AFM nanomechanics into biofilm research has fundamentally advanced our understanding of how capsular polysaccharides govern the mechanical stability and resistance of microbial communities. By bridging nanoscale cellular interactions with macroscale biofilm architecture, AFM provides unparalleled insights into the biophysical mechanisms of biofilm formation. Key takeaways include the critical role of capsular organization, influenced by structures like fimbriae, and the distinct electrokinetic signatures of polysaccharides with anti-biofilm activity. Future directions point toward the automated, large-area AFM integrated with machine learning for high-throughput analysis, and the rational design of non-biocidal anti-adhesion therapies that target the mechanical foundations of biofilm integrity. These advancements are poised to inform the next generation of biomedical interventions and clinical strategies to combat biofilm-associated antimicrobial resistance.
References
- 1. Extracellular polymeric substance [https://en.wikipedia.org/wiki/Extracellular_polymeric_substance]
- 2. Innovative Strategies Toward the Disassembly of the EPS ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2020.00952/full]
- 3. The EPS Matrix: The “House of Biofilm Cells” - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2168682/]
- 4. Bacterial Extracellular Polysaccharides in Biofilm ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4657554/]
- 5. Analysis of biofilm assembly by large area automated AFM [https://www.nature.com/articles/s41522-025-00704-y]
- 6. Multiparametric AFM reveals turgor-responsive net-like ... [https://www.nature.com/articles/ncomms8193]
- 7. Bacterial capsular polysaccharides with antibiofilm activity ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10156666/]
- 8. Recent advances in nanotechnology for eradicating bacterial ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8899562/]
- 9. Role of polysaccharides in Pseudomonas aeruginosa ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2176169/]
- 10. Studying the effect of alginate overproduction on ... [https://pubmed.ncbi.nlm.nih.gov/22121590/]
- 11. A biofilm-tropic Pseudomonas aeruginosa bacteriophage ... [https://elifesciences.org/articles/102352]
- 12. Glycoside hydrolase processing of the Pel polysaccharide ... [https://www.nature.com/articles/s41522-023-00375-7]
- 13. Modulation of virulence factors of Staphylococcus aureus ... [https://www.sciencedirect.com/science/article/pii/S0264127521004329]
- 14. Atomic Force Microscopy (AFM) on Biopolymers and ... [https://www.mdpi.com/2073-4360/14/6/1267]
- 15. Role of Capsular Polysaccharides in Biofilm Formation [https://pubmed.ncbi.nlm.nih.gov/26034816/]
- 16. Bacterial capsular polysaccharides with antibiofilm activity ... [https://www.nature.com/articles/s41467-023-37925-8]
- 17. Bacterial capsules: Occurrence, mechanism, and function [https://www.nature.com/articles/s41522-024-00497-6]
- 18. Capsular polysaccharides mimic host-tissue molecules - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4120193/]
- 19. Influence of the polysaccharide capsule on virulence and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11839663/]
- 20. Polysaccharides' Structures and Functions in Biofilm ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9965654/]
- 21. Biofilm formation: mechanistic insights and therapeutic targets [https://pmc.ncbi.nlm.nih.gov/articles/PMC10721784/]
- 22. Formation, Development, and Cross-Species Interactions ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2021.757327/full]
- 23. the polysaccharide capsule in Klebsiella is a responsive ... [https://pubs.rsc.org/en/content/articlelanding/2013/sm/c3sm51325d]
- 24. Role of Capsular Polysaccharides in Biofilm Formation: An AFM ... [https://www.semanticscholar.org/paper/2015db594fbee53b39031507ccd7bf2672582679]
- 25. Capsule Shields the Function of Short Bacterial Adhesins [https://pmc.ncbi.nlm.nih.gov/articles/PMC344426/]
- 26. 9.2: Atomic Force Microscopy (AFM) [https://chem.libretexts.org/Bookshelves/Analytical_Chemistry/Physical_Methods_in_Chemistry_and_Nano_Science_(Barron)/09%3A_Surface_Morphology_and_Structure/9.02%3A_Atomic_Force_Microscopy_(AFM)]
- 27. How to write an AFM Paper - Part 1 [https://www.afmhelp.com/index.php?option=com_content&view=article&id=145:how-to-write-an-afm-paper&catid=39&Itemid=186]
- 28. Polysaccharide functionalization reduces lipid vesicle stiffness [https://pmc.ncbi.nlm.nih.gov/articles/PMC11145274/]
- 29. Self-Assembly of Polysaccharides Gives Rise to Distinct ... [https://www.sciencedirect.com/science/article/pii/S0006349514006043]
- 30. A Practical Guide to AFM Force Spectroscopy and Data ... [https://www.bruker.com/es/products-and-solutions/microscopes/bioafm/library-assets/a-practical-guide-to-afm-force-spectroscopy-and-data-analysis.html]
- 31. How AFM Works - Atomic Force Microscopy [https://www.parksystems.com/en/learning-center/how-afm-works]
- 32. Atomic force microscopy [https://en.wikipedia.org/wiki/Atomic_force_microscopy]
- 33. AFM Principle - How Does an Atomic Force Microscope Work? [https://afm.oxinst.com/outreach/how-does-an-afm-microscope-work]
- 34. What does 2025 have in store for AFM? - NuNano [https://www.nunano.com/blog/2025/1/26/what-does-2025-have-in-store-for-afm]
- 35. Deciphering conformational dynamics in AFM data using ... [https://www.nature.com/articles/s42003-025-08365-5]
- 36. Quantitative Analysis of Amyloid-Integrated Biofilms ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3414876/]
- 37. Using Atomic Force Microscopy To Illuminate the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7447269/]
- 38. In-Situ Determination of the Mechanical Properties of Gliding ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0061663]
- 39. Quantification of bacterial adhesion forces using atomic ... [https://www.sciencedirect.com/science/article/abs/pii/S0167701299001372]
- 40. Atomic force microscopy in biofilm study [https://pubmed.ncbi.nlm.nih.gov/24793174/]
- 41. Machine learning approaches for improving atomic force ... [https://www.frontiersin.org/journals/physics/articles/10.3389/fphy.2024.1347648/full]
- 42. Analysis of biofilm assembly by large area automated AFM [https://pmc.ncbi.nlm.nih.gov/articles/PMC12062311/]
- 43. Analysis of Biofilm Assembly by Large Area Automated AFM [https://www.nanosurf.com/en/en/analysis-of-biofilm-assembly-by-large-area-automated-afm]
- 44. Large area high-speed atomic force microscopy with ... [https://www.nanopositioning.com/news/queensgate-news/large-area-high-speed-atomic-force-microscopy-with-arbitrary-scan-path-playback]
- 45. Quantitative AFM Nanomechanics Data for Machine ... [https://www.bruker.com/en/news-and-events/webinars/2021/quantitative-afm-nanomechanics-data-for-machine-learning-and-mat.html]
- 46. Relationship between Biofilm-Formation, Phenotypic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9503712/]
- 47. AFM-Based Correlative Microscopy Illuminates Human ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8138568/]
- 48. Atomic force microscopy for single molecule characterisation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6420408/]
- 49. Correlative atomic force microscopy quantitative imaging ... [https://www.nature.com/articles/s41598-018-26433-1]
- 50. A General Materials Data Science Framework for ... [https://link.springer.com/article/10.1007/s40192-024-00342-w]
- 51. A Practical Guide to AFM Force Spectroscopy and Data ... [https://www.bruker.com/it/products-and-solutions/microscopes/bioafm/library-assets/a-practical-guide-to-afm-force-spectroscopy-and-data-analysis.html]
- 52. Micro- and nano-scale adhesion of oral bacteria to ... [https://www.sciencedirect.com/science/article/pii/S1882761625000055]
- 53. Review: Advanced Atomic Force Microscopy Modes for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9775674/]
- 54. Quantitative atomic force microscopy: A statistical treatment ... [https://www.sciencedirect.com/science/article/pii/S030439912200078X]
- 55. Atomic-Force Microscopy (AFM) data: Are these images real? [https://www.openaccessgovernment.org/article/atomic-force-microscopy-afm-data-are-these-images-real/200998/]
- 56. Finite element modelling of atomic force microscopy imaging ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11580413/]
- 57. Tech Note: A Practical Guide to AFM Force Spectroscopy ... [https://www.bruker.com/en/products-and-solutions/microscopes/bioafm/resource-library/a-practical-guide-to-afm-force-spectroscopy-and-data-analysis.html]
- 58. AFM image analysis: a comprehensive guide [https://www.digitalsurf.com/blog/demystifying-afm-image-analysis-a-comprehensive-guide/]
- 59. Advances in imaging techniques for real-time microbial ... [https://www.sciencedirect.com/science/article/abs/pii/S0165993625000950]
- 60. Biofilms Exposed: Innovative Imaging and Therapeutic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12466655/]
- 61. Atomic Force Microscopy of Biofilms—Imaging, Interactions ... [https://www.intechopen.com/chapters/50739]
- 62. Atomic Force Microscopy in Microbiology: New Structural ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4120197/]
- 63. Atomic force microscopy of bacteria reveals the ... [https://www.sciencedirect.com/science/article/pii/S000527361630075X]
- 64. Diverse polysaccharide production and biofilm formation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11659406/]
- 65. Mechanisms of Multidrug Antimicrobial Resistance in Gram ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11762671/]
- 66. AFM-Based Correlative Microscopy Illuminates Human ... [https://www.frontiersin.org/journals/cellular-and-infection-microbiology/articles/10.3389/fcimb.2021.655501/full]
- 67. Quantitative Assessment of Tip Effects in Single‐Molecule ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7496922/]
- 68. Nanomaterials and Coatings for Managing Antibiotic ... [https://www.mdpi.com/2079-6382/12/2/310]
- 69. Antibiofilm polysaccharides - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3502681/]
- 70. A review of advances & potential of applying nanomaterials ... [https://www.nature.com/articles/s41545-024-00423-5]
- 71. Methods for screening and evaluation of antimicrobial activity [https://pmc.ncbi.nlm.nih.gov/articles/PMC11097785/]
- 72. analysis and critical review | npj Biofilms and Microbiomes [https://www.nature.com/articles/s41522-018-0062-5]
- 73. Microbial enzymes as powerful natural anti-biofilm candidates [https://microbialcellfactories.biomedcentral.com/articles/10.1186/s12934-024-02610-y]
- 74. Force measurements with the atomic force microscope [https://www.sciencedirect.com/science/article/abs/pii/S0167572905000488]
- 75. Atomic Force Microscopy: A Multifaceted Tool to Study ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3779510/]
- 76. Improving image contrast and material discrimination with ... [https://www.nature.com/articles/ncomms7270]