Correcting AFM Artifacts in Heterogeneous Biofilm Samples: A Guide for Reliable Nanoscale Analysis
Atomic Force Microscopy (AFM) provides unparalleled nanoscale resolution for studying biofilm structure, mechanics, and function.
Correcting AFM Artifacts in Heterogeneous Biofilm Samples: A Guide for Reliable Nanoscale Analysis
Abstract
Atomic Force Microscopy (AFM) provides unparalleled nanoscale resolution for studying biofilm structure, mechanics, and function. However, the inherent heterogeneity and soft, complex nature of biofilms introduce significant artifacts that can compromise data integrity. This article provides a comprehensive framework for researchers and drug development professionals to identify, correct, and prevent common AFM artifacts in biofilm research. Covering foundational principles, advanced methodological corrections, practical troubleshooting, and validation strategies, it synthesizes current best practices with emerging AI-driven solutions. The goal is to empower scientists to obtain more reliable, reproducible data, thereby accelerating the development of effective biofilm-control strategies in biomedical and clinical contexts.
Understanding AFM Artifacts: Why Heterogeneous Biofilms Pose Unique Imaging Challenges
Frequently Asked Questions
Q1: What are the most common categories of AFM artifacts and their primary causes? AFM artifacts can be systematically categorized based on their origin. The primary sources are the probe tip, the piezoelectric scanner, the image processing steps, and the experimental process itself [1]. The table below summarizes these categories and their key characteristics for identification.
Table: Common AFM Artifact Categories and Identification
| Artifact Category | Common Causes | Key Visual Indicators |
|---|---|---|
| Probe Artifacts [1] | Chipped tip, contaminated tip (e.g., sample debris), worn-out tip. | Repeated double features ("seeing double"), all features appear triangular or identical in size and shape, smearing of features [1]. |
| Scanner Artifacts [1] | Hysteresis, creep, non-linear piezoelectric response, poor calibration. | Image distortion, especially at the edges of the scan range; curved background; features appearing stretched or compressed [1]. |
| Image Processing Artifacts [1] | Over-aggressive line leveling, excessive filtering. | Bands across the image, unnatural smoothing, loss of fine detail, "noise nodules" from Fourier filtering [1]. |
| Process Artifacts [1] | Incorrect scanning parameters (speed, setpoint), sample contamination, external vibrations. | Features that appear or change when scanning direction is reversed, low-frequency waves, misshapen features, unusually high or low noise [1]. |
Q2: My AFM images of heterogeneous biofilms show "double" features. What is the likely cause and solution? The appearance of double or overlapping features is a classic symptom of a probe artifact, specifically a contaminated or damaged tip [1]. A contaminated tip can act as a "double tip," where both the primary tip and a piece of debris contact the sample, producing a ghost image superimposed on the real topography. To resolve this:
- Verify the artifact: Change the scan angle by 90 degrees. If the double features rotate with the scan direction, it confirms a tip issue.
- Clean the tip: Use an approved method (e.g., UV light, plasma cleaning) to remove contaminants.
- Replace the tip: If cleaning fails, the tip may be permanently damaged and must be replaced with a new, clean one [1].
Q3: I observe repeating wavy patterns in my images. Are these real sample features? Repeating, periodic waves in the background of an image are often process artifacts, not real topography. A common cause is optical interference from the AFM's laser beam [2]. This occurs when the laser reflected from the cantilever interferes with the beam reflected from the sample surface. To minimize this:
- Ensure the laser is correctly aligned and centered on the cantilever.
- Use an AFM system with a concentrated superluminescent diode (SLD) beam, which is designed to minimize such interference fringes [2].
- Check for external sources of vibration and acoustic noise.
Q4: How can I be sure my MFM signal from a biofilm is magnetic and not distorted by electrostatic forces? In Magnetic Force Microscopy (MFM), the long-range signal is a superposition of both magnetic and electrostatic forces, which can distort results [2]. This is critical for heterogeneous biofilms, which may have varying surface potentials. To isolate the true magnetic signal:
- Use a combined Kelvin Probe Force Microscopy-Magnetic Force Microscopy (KPFM-MFM) technique [2].
- This method performs real-time compensation of electrostatic interactions by applying a nullifying DC bias voltage during the second pass, leaving an artifact-free magnetic signal [2].
Troubleshooting Guide: Optimizing Scanning Parameters
Optimizing scan parameters is the first line of defense against operator-induced process artifacts. The following workflow provides a systematic approach to tuning your AFM for the best image quality on delicate biofilm samples.
Table: Key AFM Scanning Parameters and Adjustment Guidelines
| Parameter | Function | Effect of Increasing | Recommended Adjustment for Biofilms |
|---|---|---|---|
| Setpoint [1] | Defines tip-sample interaction force. | Less force, probe farther from sample. Reduces noise but may lose detail. | Start with a higher setpoint to avoid sample damage, then decrease slightly to improve detail. |
| Gain [1] | Determines control loop sensitivity. | Faster response to topography, but amplifies noise. | Increase until the system becomes unstable (chatters), then slightly decrease. |
| Scan Speed [1] | How quickly the probe rasters. | Faster scanning reduces time, but can cause smearing and loss of detail. | Use slower scan speeds for high-resolution images of soft, complex biofilm structures [1]. |
| Resolution [1] | Number of pixels in the image. | Higher resolution reveals more detail but drastically increases acquisition time. | Use a resolution (e.g., 512x512 or 1024x1024) that balances detail with acceptable scan time to minimize drift. |
Advanced Protocol: Isolating Magnetic Signals with KPFM-MFM
For researchers investigating the magnetic properties of biofilms-mineral interactions, standard MFM can be misleading. The following protocol details a method to correct for electrostatic artifacts.
Objective: To obtain an artifact-free magnetic force microscopy (MFM) signal from a heterogeneous sample by compensating for electrostatic interactions in real-time using Sideband KPFM [2].
Materials and Reagents: Table: Research Reagent Solutions for KPFM-MFM
| Item | Function / Specification |
|---|---|
| Non-Magnetic Sample Holder | Prevents magnetic signal interference from the holder itself [2]. |
| Cobalt Alloy Coated Tip (e.g., PPP-MFMR) | Provides sensitivity to both magnetic and electrostatic forces [2]. |
| AFM with Multiple Lock-in Amplifiers | Essential for simultaneous topography, KPFM, and MFM signal detection (e.g., Park FX40) [2]. |
Methodology:
- Sample Preparation: Mount the sample on a non-magnetic holder to exclude external magnetic interference [2].
- First Pass (Topography):
- Engage the tip in non-contact mode.
- Use the first lock-in amplifier exclusively for topography feedback to record the true surface profile [2].
- Second Pass (KPFM-MFM):
- Lift the tip to a constant height (e.g., 50-500 nm) above the previously recorded topography.
- Simultaneously perform the following:
- Sideband KPFM: Apply an AC voltage (ωAC) to the tip. Use the second and third lock-in amplifiers to detect the sidebands (ωAC+ω₀ and ωAC-ω₀) and calculate the contact potential difference (CPD). Apply a nullifying DC bias (VDC) to cancel the electrostatic force [2].
- MFM: Use the amplitude or phase shift from the first lock-in amplifier to detect the magnetic force, which is now isolated after electrostatic compensation [2].
The logical relationship and signal flow of this advanced technique are illustrated below.
Atomic Force Microscopy (AFM) is a powerful tool for studying biofilms, providing high-resolution topographical images and quantitative maps of nanomechanical properties under physiological conditions without extensive sample preparation [3]. However, the inherent complexity of biofilms—characterized by their heterogeneous structure, the presence of extracellular polymeric substances (EPS), and varied cellular morphology—makes them particularly susceptible to imaging artifacts. These artifacts can distort data and lead to incorrect interpretations of biofilm architecture and properties. This guide provides researchers with practical troubleshooting advice to identify, mitigate, and correct common AFM artifacts in biofilm research.
Troubleshooting Guide: Identifying and Resolving Common AFM Artifacts
FAQ: What are the most frequent AFM artifacts when imaging biofilms, and how can I fix them?
Q1: My biofilm images show strange, repeating patterns or features that look duplicated. What is the cause? This is a classic tip artifact [4] [5]. It occurs when the AFM tip is damaged, contaminated with debris from the biofilm, or is a "double tip" [5]. A contaminated or blunt tip will produce images where features appear larger than they are, trenches seem smaller, and irregular shapes repeat across the scan [4].
- Solution: Replace the AFM probe with a new, clean one. To minimize contamination, ensure your sample preparation protocols minimize loosely adhered material [4]. For biofilms, using conical tips instead of pyramidal ones can also help reduce artifact issues [4].
Q2: I am seeing repetitive horizontal lines across my entire image. What could be causing this? This is typically caused by electrical noise or laser interference [4].
- Solution:
- For Electrical Noise (50 Hz): This is often governed by the building's electrical circuits. You can try imaging during quieter periods (e.g., early mornings or late evenings) when electrical noise is reduced [4].
- For Laser Interference: This happens with reflective samples; laser light reflects off the sample and interferes with the signal. Using a probe with a reflective coating (e.g., gold or aluminium) on the cantilever can eliminate this problem [4].
Q3: I see streaks or oscillations in my images, especially on rough areas of the biofilm. Why? This is often due to environmental noise/vibration or surface contamination [4] [5]. Biofilms are often imaged in liquid, and vibrations from doors, traffic, or people can easily disrupt the scan. Furthermore, loose EPS or cells on the surface can be pushed by the tip, creating streaks.
- Solution: Ensure the AFM's anti-vibration table is functioning. Place a "STOP AFM in Progress" sign to alert others. Image during quiet hours or relocate the instrument to a quieter room [4]. For contamination, refine sample preparation to remove unattached cells and debris gently but thoroughly [3] [4].
Q4: My image is distorted, with features that seem stretched or skewed. This is likely caused by sample drift or a piezo scanner artifact known as piezo creep [5]. Sample drift can occur if the biofilm is not securely fixed. Piezo creep is an inherent property of the piezoelectric scanner material.
- Solution: Allow sufficient time for the AFM and sample to thermally equilibrate after loading. For scanner artifacts, most modern AFM software includes algorithms to correct for these distortions. Ensure your system's calibration is up to date.
Quantitative Data on Bacterial Adhesion Forces
AFM can quantify interaction forces at the nanoscale, which is crucial for understanding initial biofilm attachment. The table below summarizes typical adhesion forces measured on a single bacterium, highlighting how forces vary at different locations due to EPS accumulation.
Table 1: Quantified Adhesion Forces in Bacterial Systems Measured by AFM
| Measurement Location | Adhesion Force Range (nN) | Probable Cause |
|---|---|---|
| AFM Tip vs. Bacterial Cell Surface | -3.9 to -4.3 nN | Direct tip-cell surface interaction [6] |
| Periphery of Cell-Substratum Contact | -5.1 to -5.9 nN | Accumulation of EPS at the attachment point [6] |
| Cell-Cell Interface | -6.5 to -6.8 nN | Significant EPS bridging between adjacent cells [6] |
Advanced Method: Large-Area AFM and Machine Learning
A major challenge in biofilm research is the scale mismatch between AFM's small imaging area (typically <100 µm) and the millimeter-scale heterogeneity of biofilms [3]. An advanced solution is an automated large-area AFM approach, which captures high-resolution images over millimeter-scale areas.
- Experimental Protocol:
- Sample Preparation: A Petri dish containing PFOTS-treated glass coverslips is inoculated with the bacterial strain (e.g., Pantoea sp. YR343). At selected time points, a coverslip is removed, gently rinsed to remove unattached cells, and dried before imaging [3].
- Automated Scanning: The AFM is programmed to automatically collect multiple adjacent high-resolution images across a large area of the sample.
- Image Stitching: Machine learning (ML) algorithms stitch the individual images together into a seamless, high-resolution mosaic with minimal overlap for speed [3].
- Data Analysis: ML-based image segmentation and analysis tools automatically extract quantitative parameters such as cell count, confluency, cell shape, and orientation from the large-area dataset [3].
This methodology overcomes the limitation of small scan areas and provides a comprehensive view of spatial heterogeneity, revealing patterns like a honeycomb structure of surface-attached cells that were previously obscured [3].
Visual Guide to Troubleshooting AFM Artifacts in Biofilm Research
The following diagram illustrates a systematic workflow for diagnosing and resolving the most common AFM artifacts encountered when imaging biofilms.
Diagram 1: AFM Artifact Troubleshooting Guide
The Scientist's Toolkit: Key Reagents and Materials for AFM Biofilm Studies
Table 2: Essential Research Reagents and Materials for AFM Biofilm Experiments
| Item | Function in AFM Biofilm Research | Example from Literature |
|---|---|---|
| PFOTS-treated Glass | Creates a hydrophobic surface to study specific bacterial attachment dynamics and early biofilm formation [3]. | Used to examine the organization of Pantoea sp. YR343, revealing a preferred cellular orientation [3]. |
| Freshly Cleaved Mica | Provides an atomically flat, clean substrate for depositing and immobilizing bacterial cells or particles for high-resolution imaging [6]. | Used as a substrate for imaging sulfate-reducing bacteria (SRB) to quantify adhesion forces [6]. |
| High Aspect Ratio (HAR) Conical Probes | Superior for accurately resolving steep-edged features and deep trenches in heterogeneous biofilm structures, minimizing tip-convolution artifacts [4]. | Recommended over pyramidal tips for imaging high aspect ratio features common in biofilms [4]. |
| Reflectively Coated Cantilevers | Metal coatings (e.g., gold, aluminium) prevent laser interference artifacts, which are common when scanning reflective surfaces or in liquid [4]. | Coating prevents interference from laser light reflecting off the sample or through a semi-transparent cantilever [4]. |
| Modified Postgate's Medium C | A specific growth medium used for cultivating sulfate-reducing bacteria (SRB) isolated from environments like marine sediments [6]. | Used to culture SRB for AFM studies on adhesion and biofilm formation on mica surfaces [6]. |
Atomic Force Microscopy (AFM) provides high-resolution, three-dimensional topographical images of biofilms, which are complex microbial communities critical in medical, industrial, and environmental contexts. Unlike electron microscopy, AFM can image samples under physiological conditions with minimal preparation, enabling researchers to visualize native biofilm structures, including extracellular polymeric substances (EPS), flagella, and individual microbial cells. However, when imaging heterogeneous biofilm samples, several artifacts can compromise data accuracy. These include distortions from tip convolution, image deformation from scanner drift, and measurement errors from adhesion forces. Understanding, identifying, and correcting these artifacts is essential for obtaining reliable, high-quality data in biofilm research and drug development applications. This guide provides troubleshooting protocols to address these common challenges.
Troubleshooting FAQs and Guides
FAQ: Common AFM Artifacts and Solutions
Q1: Why do my biofilm images show repeated, irregular patterns or features that look duplicated? A: This is typically a tip artifact. A contaminated or broken probe tip interacts with the sample surface in a way that distorts the true topography. With a blunt tip, structures appear larger, and trenches appear smaller. This can obscure critical biofilm features like the width of bacterial flagella or the shape of pores in the EPS matrix [4].
- Solution: Replace the AFM probe with a new, sharp one. For consistent high-resolution imaging of biofilms, use probes with a high aspect ratio and a sharp apex. Ensure proper sample preparation to minimize loose debris that could contaminate the tip [4].
Q2: Why are my AFM images of a bacterial cluster distorted or stretched in one direction? A: This is characteristic of sample drift. The sample moves slowly in one direction during the scan, often due to thermal expansion as the instrument settles. This is particularly problematic in high-resolution techniques like AFM and makes it difficult to accurately measure cellular dimensions and distances between cells in a biofilm [7].
- Solution: Allow the system sufficient time to equilibrate thermally after loading the sample. Turn off external heat sources (e.g., lights) and ensure the sample is securely fixed. Scanning quickly can also help reduce the visible effects of drift [7].
Q3: Why am I having difficulty accurately imaging deep trenches or vertical structures in my multilayer biofilm? A: This is often due to the geometry of the AFM probe. Pyramidal or tetrahedral tips have side-walls that can make physical contact with steep-edged features before the tip apex reaches the bottom of a trench. Furthermore, low aspect ratio probes cannot physically reach into deep and narrow pores within the biofilm structure [4].
- Solution: Switch to a probe with a conical shape and a high aspect ratio (HAR). Conical tips trace steep edges more accurately, and HAR probes are specifically designed to resolve high, non-planar features common in complex biofilms [4].
Q4: Why do I see repetitive lines or streaks across my image? A: This is usually caused by external interference.
- Electrical noise from building circuits or other instruments appears as periodic lines at 50/60 Hz. You can identify it by comparing the noise frequency to your scan rate [4].
- Laser interference occurs when light reflects off a highly reflective sample or the cantilever itself, creating interfering signals at the photodetector [4].
- Environmental vibrations from doors, people, or traffic can also cause streaking [4].
- Solution: For electrical noise, try imaging during quieter periods (e.g., early morning). Use a probe with a reflective coating to minimize laser interference. Ensure the anti-vibration table is functional and place the AFM in a quiet location, using signs to alert others of sensitive work in progress [4].
Troubleshooting Guide: Step-by-Step Protocols
Protocol 1: Correcting for Tip Convolution Artifacts
Tip convolution is a fundamental artifact where the finite size and shape of the AFM tip physically interacts with the sample, causing steep features to appear broader and shallower than they are. This is critical when imaging nanoscale features like flagella (~20-50 nm in height) [3] or pore spaces in the EPS.
- Step 1: Identify the Artifact. Look for features that seem unusually broadened or repeated across the image. Fine details may be lost or appear with unrealistic symmetry.
- Step 2: Characterize the Tip Geometry.
- Method A (Blind Estimation): Use a software algorithm (e.g., in Gwyddion) with your image data from a sample with sharp features to estimate the actual tip shape [8].
- Method B (Modeling): If the tip is new and clean, use the manufacturer's specifications (apex radius, slope) to create a tip model in your analysis software [8].
- Step 3: Reconstruct the Surface.
- Use a surface reconstruction (erosion) algorithm in your analysis software. This process mathematically "erodes" the scanned image with the known tip shape to produce a closer approximation of the true surface [8].
- Step 4: Generate a Certainty Map.
- After reconstruction, create a certainty map. This highlights areas of the image where the tip did not make single-point contact with the surface and where data is irreversibly lost, helping you assess the reliability of the corrected image [8].
Table 1: Quantitative Impact of Tip Convolution on Biofilm Features
| Biofilm Feature | Actual Size (approx.) | Apparent Size with Blunt Tip | Correction Method |
|---|---|---|---|
| Bacterial Flagella | 20-50 nm height [3] | Broader, may not be resolved | Tip reconstruction, sharper probes |
| EPS Fibrils | 10-100 nm diameter | Appear thicker | Tip reconstruction, certainty mapping |
| Pores in EPS Matrix | Variable, can be <100 nm | Appear narrower and shallower | Use of high-aspect-ratio probes [4] |
| Single Bacterial Cell | ~2 µm length, ~1 µm diam. [3] | Dimensions slightly enlarged | Surface reconstruction algorithm [8] |
Protocol 2: Mitigating Sample Drift in Long-Duration Biofilm Scans
Sample drift is a major concern for time-lapse studies of biofilm growth or for large-area scans that take a long time, as it distorts spatial relationships.
- Step 1: Recognition. Drift is confirmed when the same cluster of cells appears distorted and different when the slow scan direction is changed [7].
- Step 2: System Stabilization.
- Thermal Equilibrium: Load the sample and allow the AFM stage to sit for at least 30-60 minutes before starting a high-resolution or long-duration scan. This allows thermal equilibration to minimize drift from expansion/contraction.
- Secure Mounting: Ensure the biofilm substrate (e.g., a glass coverslip or pyrite coupon [9]) is firmly fixed to the sample stage using a reliable adhesive or clip.
- Environmental Control: Turn off nearby heat sources and maintain a stable room temperature.
- Step 3: Scanning Parameter Adjustment.
- Increase the scan speed to "outrun" the drift, though this must be balanced against maintaining image quality and minimizing tip forces. For large-area automated AFM, software with automated drift compensation may be available [3].
Table 2: Common Artifacts and Their Impact on Biofilm Analysis
| Artifact Type | Effect on Biofilm Image | Impact on Quantitative Analysis |
|---|---|---|
| Tip Convolution | Loss of fine detail, broadening of nanofeatures, inaccurate trench profiles | Inaccurate measurement of flagellar diameter, EPS fiber size, and surface porosity [8] |
| Sample Drift | Stretching or compression of features in one direction, distorted cell clusters | Incorrect calculation of cell-to-cell distances, cluster size, and spatial distribution [7] |
| Adhesion Forces | Sudden jumps in height data, "ghost" features, difficulty maintaining setpoint | Inaccurate nanomechanical property mapping (e.g., adhesion, stiffness) [4] |
| Electrical Noise | Repetitive horizontal lines across the image | Reduced signal-to-noise ratio, obscuring subtle topographical changes |
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for AFM Biofilm Experiments
| Item | Function in Biofilm AFM | Example & Notes |
|---|---|---|
| High-Aspect-Ratio (HAR) Probes | To accurately resolve deep trenches and vertical structures in 3D biofilm architectures. | Conical silicon or silicon nitride tips; superior to pyramidal tips for non-planar features [4]. |
| Sharp Probes (High Resolution) | For imaging nanoscale features like flagella, pili, and EPS fibers. | Probes with a low tip radius (<10 nm) are essential for high-resolution scans [3] [4]. |
| Chemically Functionalized Probes | To measure specific adhesion forces between the tip and biofilm components. | Tips coated with specific molecules (e.g., lectins for polysaccharide binding) can map interaction forces. |
| Opaque & Reflective Substrates | For combined AFM-Epifluorescence microscopy on non-transparent materials. | Pyrite coupons for bioleaching studies [9]; reflective coatings can reduce laser interference [4]. |
| Surface Treatment Reagents | To study the effect of surface properties on bacterial adhesion and biofilm assembly. | PFOTS-treated glass to create hydrophobic surfaces [3]. |
| Fluorescent Stains (e.g., DAPI) | For correlative microscopy; allows identification of cellular features in AFM topographs. | Stains bacterial DNA, enabling cell identification on opaque surfaces when combined with EFM [9]. |
Experimental Protocols from Key Studies
Detailed Methodology: Combined AFM and Epifluorescence Microscopy (EFM) for Biofilms on Opaque Surfaces
This protocol, adapted from a study on Acidithiobacillus ferrooxidans biofilms on pyrite, is ideal for correlating topographic data with cell identity on opaque substrates common in biocorrosion and bioleaching research [9].
Sample Preparation:
- Substratum: Cut coupons (e.g., 10x10x1 mm) from an opaque material of interest (e.g., pyrite, metal alloy). Clean thoroughly (e.g., with HCl and acetone for pyrite) to remove contaminants [9].
- Biofilm Growth: Incubate sterile coupons in a bacterial suspension for the desired attachment and biofilm formation period (e.g., 4 days at 28°C) [9].
- Staining: After incubation, stain the biofilm with a fluorescent dye (e.g., 0.01% DAPI for 10 minutes) to label cellular components. The sample can be either kept hydrated or air-dried [9].
Instrumentation and Shuttling:
- Use a shuttle stage system that allows precise transfer of the sample between a separate AFM and an upright epifluorescence microscope.
- Fix the stained and dried sample to a glass slide and mount it on the shuttle stage.
Correlative Imaging:
- EFM Imaging: First, locate an area of interest and capture a fluorescence image using the EFM to identify the positions of bacterial cells.
- AFM Imaging: Transfer the shuttle stage to the AFM. Navigate to the exact same location (with an error of ~3-5 µm) using the predefined coordinates. Acquire high-resolution topographical images in air or liquid. Note that contact mode in liquid may deform or detach cells, while imaging in air is often more stable [9].
Data Correlation: Overlay the EFM and AFM images to correlate cell identity (from fluorescence) with high-resolution topography and mechanical properties (from AFM).
Combined AFM-EFM Workflow
Advanced Methods: Large-Area AFM and Machine Learning
Traditional AFM is limited to scan areas typically below 100x100 µm, making it difficult to link cellular-scale events to the larger functional architecture of biofilms [3]. Recent advancements have begun to overcome this limitation.
Automated Large-Area AFM with Machine Learning: A novel approach involves automating the AFM to capture and stitch together multiple high-resolution images over millimeter-scale areas [3]. This process is aided by machine learning (ML) for several key tasks:
- Image Stitching: Seamlessly combines adjacent image tiles, even with minimal overlapping features.
- Cell Detection and Classification: Automatically identifies and categorizes individual cells within the vast dataset, extracting parameters like cell count, confluency, shape, and orientation [3].
- Application: This method has revealed a preferred cellular orientation and a distinctive honeycomb pattern in early-stage biofilms of Pantoea sp. YR343, features previously obscured by the limited field of view [3].
Resolving Scale Mismatch with AFM
Atomic Force Microscopy (AFM) is a powerful tool for characterizing the structural and mechanical properties of heterogeneous biofilms at the nanoscale. However, the inherent complexity of biofilms—with their varied cellular morphologies, extracellular polymeric substances (EPS), and intricate surface topography—makes them particularly susceptible to imaging and measurement artifacts. These artifacts can significantly skew topographical data and nanomechanical properties, leading to erroneous biological interpretations. This technical support guide provides a systematic framework for identifying, troubleshooting, and correcting common AFM artifacts within the context of biofilm research, ensuring data integrity for critical applications in drug development and microbial science.
Frequently Asked Questions (FAQs) & Troubleshooting Guides
FAQ 1: My biofilm images show repeated, "ghosted" features or irregular shapes that do not align with expected cellular structures. What is the cause and how can I fix it?
- Problem: Unexpected patterns in images, such as duplicated features or irregular shapes repeating across the scan [4].
- Primary Cause: Tip artifacts caused by a contaminated, damaged, or blunt AFM probe [4] [5]. A dirty or broken tip interacts with the sample in an unpredictable way, creating features that are not real.
- Solutions:
- Replace the Probe: The most straightforward solution is to use a new, sharp probe [4].
- Verify Probe Quality: Ensure you are using a probe appropriate for your sample. Contaminated probes can often be cleaned, but replacement is more reliable.
- Sample Preparation: Minimize loosely adhered material on your sample surface to reduce the chance of contamination transfer to the tip [4].
- Problem: Inaccurate profiling of vertical structures and deep trenches [4] [10].
- Primary Cause: Tip convolution effects [10] [5]. This occurs when the geometry and finite size of the AFM tip prevent it from reaching the bottom of narrow features, leading to a distorted image where features appear wider and trenches appear narrower [4].
- Solutions:
- Use High-Aspect-Ratio (HAR) Probes: HAR probes are designed to penetrate deep, narrow features more effectively, providing a more accurate topographic profile [4].
- Use Conical Tips: For features with high vertical relief, conical tips are superior to standard pyramidal or tetrahedral tips as they provide better access to steep-edged structures [4].
- Tip Deconvolution: Apply post-processing algorithms to mathematically correct for the known geometry of the tip [10].
FAQ 3: Repetitive horizontal lines or streaking appear across my images, obscuring the true biofilm topography.
- Problem: Repetitive lines or streaks in the fast-scan direction [4] [1].
- Primary Causes and Solutions:
- Electrical Noise: This often manifests as 50/60 Hz interference. To fix it, try changing the scan rate or identify times of day with lower electrical load on the building's circuits [4].
- Laser Interference: If the sample is highly reflective, laser light reflecting off the sample surface can interfere with the signal. Use a probe with a reflective coating on the cantilever to mitigate this [4].
- Environmental Vibration: Noise from building vibrations, doors, or traffic can cause streaking. Ensure the anti-vibration table is functional and consider imaging during quieter periods or relocating the instrument to a basement lab [4].
- Scanning Too Fast: If the scan speed is too high, the feedback loop cannot track the surface accurately, causing streaks. Reduce the scan rate to improve image quality [1].
FAQ 4: The measured Young's modulus of individual collagen fibrils or other nanofibers in my biofilm matrix shows an unacceptably wide range of values. What are the potential sources of error?
- Problem: High variability and inaccuracy in nanomechanical measurements of fibrous structures [10].
- Primary Causes:
- Invalid Elastic Half-Space Assumption: The standard Hertzian contact models used for calculating Young's modulus assume the sample is an infinitely thick, flat, elastic half-space. This assumption is invalid for nanofibers, where the radius is comparable to the tip radius [10].
- Tip Convolution in Radius Measurement: The accurate determination of the fibril's radius is critical for mechanics models. Tip convolution effects lead to an overestimation of the fibril's width, which directly propagates into an error in the calculated Young's modulus [10].
- Incorrect Contact Model: Using a model for a perfect conical or pyramidal indenter when the actual tip shape is different or poorly characterized introduces significant error [10].
- Solutions:
- Apply Correction Factors: Use adjusted equations from Hertzian mechanics that incorporate "correction factors" for the finite dimensions of the nanofiber [10].
- Accurate Tip Characterization: Precisely determine the shape and dimensions of the AFM tip through calibration samples to correctly model the contact area [10].
- Validate Radius Measurement: Use techniques like scanning electron microscopy (SEM) to independently verify the nanofiber radius, avoiding errors from AFM tip convolution [10].
The following tables summarize the common artifacts, their impact on quantitative data, and the corresponding solutions for biofilm research.
Table 1: Impact of Common Artifacts on Topographical and Mechanical Data
| Artifact Type | Effect on Topography | Effect on Nanomechanics | Common in Biofilm Features |
|---|---|---|---|
| Tip Convolution [10] | - Overestimation of feature width [10]- Underestimation of trench depth [4] | - Invalidates mechanical models [10]- Incorrect contact area calculation [10] | Bacterial cells [3], EPS fibrils [10], pore networks |
| Contaminated Tip [4] [5] | - "Double-tip" ghost images [5]- Strange, non-reproducible shapes [4] | - Unreliable force-distance curves- Spurious adhesion & stiffness values | All surfaces, especially with loose EPS [4] |
| Blunt Tip [4] | - Loss of high-resolution details- Features appear smeared and larger | - Overestimation of modulus (larger contact area)- Poor spatial resolution in property mapping | Flagella [3], surface proteins [3], fine EPS structures |
| Electrical/Environmental Noise [4] [1] | - Repetitive stripes/streaks in image- Increased background noise | - Noisy force spectroscopy data- Reduced accuracy in fitting models | All measurements, particularly high-resolution scans |
Table 2: Research Reagent Solutions for AFM Biofilm Characterization
| Essential Material / Reagent | Function and Application | Considerations for Biofilm Research |
|---|---|---|
| High-Aspect-Ratio (HAR) Probes [4] | Provides accurate topography of deep, narrow features like EPS pores and bacterial cell junctions. | Superior for resolving the complex 3D architecture of heterogeneous biofilms. |
| Conical Tips [4] | Improves profiling of features with steep edges, such as bacterial clusters and biofilm aggregates. | Reduces tip convolution artifacts on complex biofilm surfaces. |
| Reflective Coated Cantilevers [4] | (e.g., Gold, Aluminum) Minimizes laser interference artifacts on reflective substrates. | Essential for imaging biofilms on abiotic surfaces like medical implants or silicon wafers. |
| PFOTS-treated Glass Surfaces [3] | Creates a defined hydrophobic surface to study initial bacterial attachment and biofilm assembly. | Useful for standardizing adhesion studies across experiments. |
Experimental Protocol: An Integrated Workflow for Artifact Correction in Biofilm AFM
This detailed protocol outlines a systematic approach for acquiring and verifying artifact-free AFM data from biofilm samples, integrating both operational and computational steps.
Step 1: Pre-Imaging Preparation and Probe Selection
- Sample Preparation: Gently rinse the biofilm to remove unattached cells and culture medium, but avoid dehydration if measuring under physiological conditions [3]. For biofilm studies on abiotic surfaces, treatments like PFOTS can standardize surface properties [3].
- Probe Selection: Choose a probe based on the biofilm's key features.
- For high-resolution imaging of flagella or fine EPS, use sharp tips (nominal radius < 10 nm) [3].
- For mapping the topography of complex, clustered biofilms, use High-Aspect-Ratio (HAR) or conical tips to minimize convolution [4] [10].
- For nanomechanical mapping, select a tip with a well-defined geometry (e.g., spherical colloidal probes) and a spring constant appropriate for the expected stiffness of the biofilm.
Step 2: Initial Setup and In-Run Optimization
- Scanner Calibration: Use a calibration grating with known dimensions to verify the accuracy of the scanner in X, Y, and Z axes. This corrects for scanner artifacts like hysteresis and creep [1].
- Laser Alignment: Carefully align the laser on the cantilever to prevent interference patterns caused by reflections from the sample surface [4] [1].
- Parameter Tuning:
- Start by increasing the gain to improve the feedback response [1].
- Then, decrease the setpoint to increase the tip-sample interaction for better tracking, but ensure the force is low enough to avoid sample damage [1].
- Use a slow scan speed to allow the tip to accurately track the heterogeneous and soft biofilm surface. Increase speed only if the image quality remains high [1].
Step 3: Post-Processing and Data Validation
- Tip Deconvolution: If the tip shape is well-characterized, apply deconvolution algorithms to the topographical data to correct for broadening effects, providing a more accurate representation of feature dimensions [10].
- Model Selection for Nanomechanics: For mechanical data on fibrous structures (e.g., collagen in EPS), do not use standard Hertz models. Apply corrected models that account for the cylindrical geometry of the fibril and the relative dimensions of the tip and sample [10].
- Data Cross-Validation: Correlate AFM data with other imaging modalities. For instance, use Confocal Laser Scanning Microscopy (CLSM) to validate the overall biofilm architecture or SEM to verify the dimensions of specific features [3] [11].
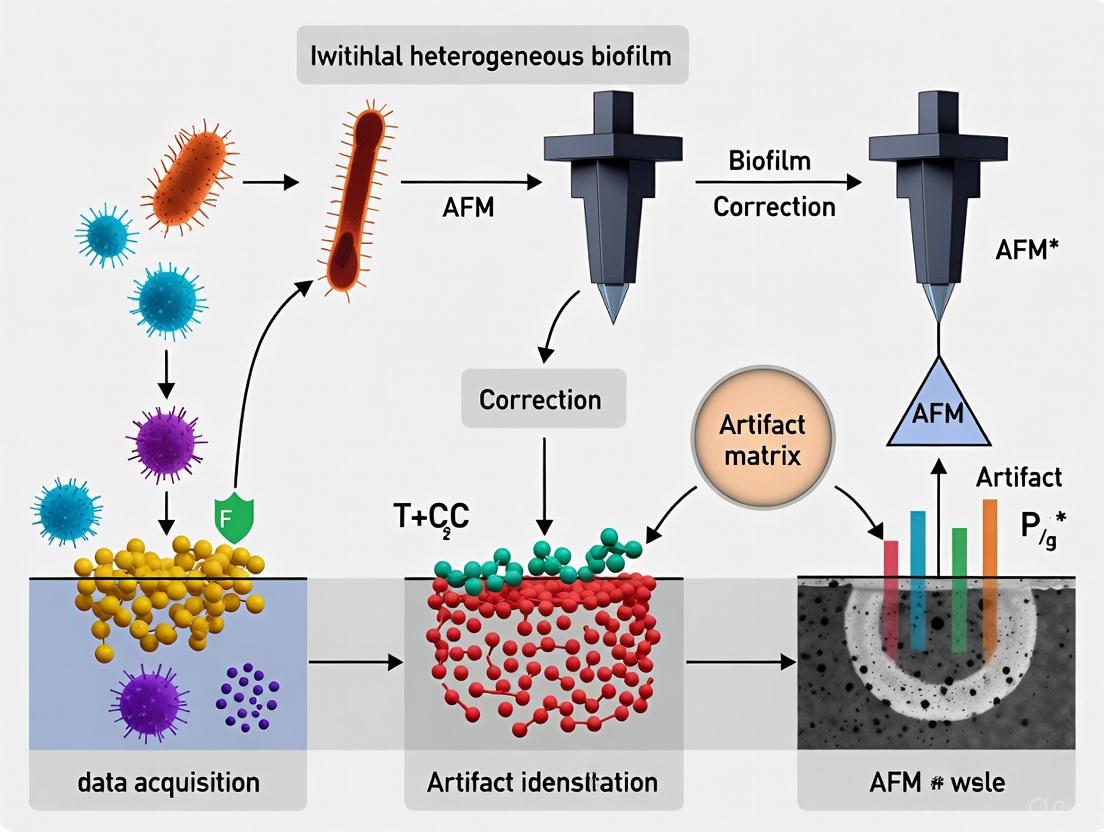
Workflow Diagram: An AI-Enhanced Framework for Artifact Management
The following diagram illustrates a modern, integrated workflow that combines traditional AFM best practices with machine learning (ML) and artificial intelligence (AI) approaches to proactively manage artifacts in biofilm research, as highlighted in recent literature [3] [11].
Diagram Title: Integrated AFM Workflow for Biofilm Artifact Management
This workflow highlights how traditional practices are augmented by AI. For example, automated large-area AFM combined with machine learning-based stitching allows for the creation of high-resolution millimeter-scale maps, revealing biofilm patterns like honeycomb structures previously obscured by small scan areas [3]. Furthermore, AI-powered segmentation can automatically detect cells, classify features, and identify regions likely affected by artifacts, guiding the researcher to areas requiring re-scanning or specific correction protocols [3] [11]. This integrated approach significantly enhances the throughput, reliability, and depth of AFM analysis in complex biofilm systems.
Advanced AFM Methodologies and Protocols for Artifact Minimization in Biofilms
Atomic Force Microscopy (AFM) generates nanoscale topographical and mechanical data by physically scanning a sharp probe across a sample surface. In the context of heterogeneous biofilm research, an inappropriate probe choice is a primary source of imaging artifacts and inaccurate nanomechanical data. Biofilms present a unique challenge due to their complex architecture, combining soft, adhesive extracellular polymeric substances (EPS) with intricate, high-aspect-ratio features like pores, channels, and cellular aggregates [12] [13]. This technical guide outlines evidence-based procedures for selecting optimal AFM probes to accurately characterize biofilm systems, thereby correcting common artifacts and ensuring data fidelity for researchers and drug development professionals.
Key Probe Parameters and Their Impact on Biofilm Imaging
The AFM probe is a complex tool whose components significantly influence data quality on heterogeneous soft materials [14] [13]. The following parameters are most critical for biofilm characterization.
Critical Probe Selection Parameters
- Force Constant: This defines the stiffness of the probe cantilever. For soft, adhesive samples like biofilms, a probe with a low to moderate force constant (typically 0.1 N/m to 5 N/m) is recommended [14]. A stiffer probe can damage soft samples, while a more flexible cantilever is sensitive enough to track the surface without excessive force. If the biofilm is particularly sticky, a slightly stiffer probe (e.g., towards the higher end of this range) can help the cantilever break away from adhesive interactions during oscillatory modes like tapping mode [14].
- Resonant Frequency: In dynamic (tapping) modes, the probe is oscillated near its resonant frequency. A higher resonant frequency (generally >300 kHz) is desirable because it allows the probe's tapping frequency to be far from the lower scanning frequency. This separation makes it easier to isolate the sample's topographical response and reduces the chance of feedback instability and associated artifacts [14].
- Tip Radius: The sharpness of the tip dictates the best achievable lateral resolution. A sharp tip radius (<10 nm) is necessary to resolve fine features such as individual flagella (which can be 20-50 nm in height) or pores within the EPS matrix [12] [14]. A tip radius larger than the feature of interest will result in broadening or complete obscuration of that feature.
- Tip Aspect Ratio and Shape: For biofilms with deep crevices or high-aspect-ratio features, a high-aspect-ratio tip (tall and skinny) is essential. Standard pyramidal tips cannot probe deep into indentations, leading to "shadowing" or "widening" artifacts. Specialized tip geometries (e.g., needle-like) are designed for this purpose [14].
- Tip Coating: Tips with hard, durable coatings (e.g., diamond-like carbon) can prolong probe lifetime when scanning rough or abrasive samples. However, coatings can increase the tip radius, so a balance must be struck between longevity and ultimate resolution [14].
Table 1: AFM Probe Parameter Guide for Biofilm Characterization
| Parameter | Recommendation for Biofilms | Rationale | Risk of Incorrect Selection |
|---|---|---|---|
| Force Constant | 0.1 - 5 N/m | Provides sufficient sensitivity for soft samples while overcoming adhesion. | Too stiff: sample damage; Too soft: poor tracking, stickiness |
| Resonant Frequency | >300 kHz | Isolates tapping frequency from scan frequency, improving stability. | Low frequency: increased artifacts, slower scan speeds |
| Tip Radius | <10 nm | Enables resolution of fine features (e.g., flagella, EPS fibers). | Large radius: poor resolution, feature broadening |
| Tip Aspect Ratio | High | Allows probing into deep, narrow surface features. | Low aspect ratio: shadowing, inaccurate depth measurement |
| Q Factor | High (for rectangular levers) | Increases sensitivity to small force variations. | Low Q: reduced signal-to-noise ratio |
Troubleshooting Guide: Common Probe-Related Artifacts and Solutions
This section addresses specific issues users might encounter during AFM experiments on biofilm samples.
FAQ 1: My images of a bacterial cluster show a "double-tip" or "ghost" artifact. What is the cause and how can I fix it?
- Problem: A "double-tip" artifact, where features appear to be duplicated, indicates a damaged or contaminated probe tip.
- Solution:
- Inspect the tip: Use a high-resolution optical microscope or SEM if available to confirm tip integrity.
- Clean the sample and tip: Ensure your biofilm sample is free of loose debris. In some cases, tip cleaning procedures can remove contaminants.
- Replace the probe: If the tip is physically broken, replacement is the only solution. Handle probes with care using an ESD bracelet to prevent electrostatic discharge damage [14].
FAQ 2: When scanning a heterogeneous biofilm, my probe appears to "get stuck" in soft areas, distorting the image.
- Problem: This is a classic sign of excessive adhesion forces between the probe and the soft, adhesive biofilm matrix.
- Solution:
- Increase the cantilever stiffness: Switch to a probe with a higher force constant (e.g., 2-5 N/m) to provide enough restoring force to pull away from the sticky surface [14].
- Optimize scanning parameters: Reduce the scan size and/or increase the scan rate to spend less time in contact with the adhesive area.
- Use dynamic mode: If using a contact mode, switch to a dynamic (tapping) mode, which minimizes lateral forces and adhesion effects [13].
FAQ 3: I cannot resolve the fine, web-like structures (like flagella) between cells that I know are present.
- Problem: This is a resolution limitation, typically caused by a tip that is too blunt or has an inappropriate shape.
- Solution:
- Use a sharper tip: Select a probe with a guaranteed tip radius of <5 nm.
- Select a high-aspect-ratio tip: For features that extend from the surface, a tall, sharp tip is necessary to accurately trace them without convolution from the tip shank [14].
- Verify with a reference sample: Image a known sample with sharp features (e.g., a grating) to confirm your probe's effective sharpness.
FAQ 4: My force spectroscopy data on a biofilm shows an artificially high modulus near the cell-EPS boundary. Why?
- Problem: This is likely a substrate effect or convolution artifact. The probe's interaction volume includes both the soft EPS and the stiffer underlying cell or substrate, convoluting the measurement [15] [13].
- Solution:
- Use a colloidal probe: A spherical tip provides a well-defined contact geometry that is easier to model and less sensitive to sharp transitions.
- Apply advanced contact mechanics: Use models that account for multi-layer systems or finite sample thickness.
- Shallow indentation: Use very low indentation depths relative to the feature size to locally probe the soft material, though this requires high sensitivity [13].
Experimental Protocol: Correlating Probe Selection with Biofilm Feature Analysis
The following methodology, adapted from recent high-impact research, provides a workflow for selecting and applying AFM probes to characterize key features in a Pantoea sp. biofilm.
Title: Protocol for High-Resolution Topographical and Nanomechanical Mapping of Early-Stage Biofilms
Background: This protocol is designed to capture the spatial heterogeneity and cellular morphology during the early stages of biofilm formation, which includes imaging delicate extracellular structures and measuring local mechanical properties [12].
Materials and Reagents:
- Biofilm Sample: Pantoea sp. YR343 biofilm grown on PFOTS-treated glass coverslips for ~30 minutes to 8 hours [12].
- Imaging Buffer: Appropriate liquid growth medium to maintain physiological conditions.
- AFM Probes: See "Research Reagent Solutions" below.
Procedure:
- Sample Preparation: Gently rinse the coverslip with buffer to remove unattached cells. If using liquid imaging, mount the sample in the fluid cell.
- Probe Selection: Based on the target feature, select the appropriate probe (see Table 2).
- AFM Calibration: Precisely calibrate the AFM's photodetector, cantilever sensitivity, and spring constant using a standardized method (e.g., thermal tune) on a clean, rigid surface (e.g., silicon wafer) [15].
- Coarse Approach: Carefully engage the probe onto the sample surface in a region with low feature density.
- Topographical Imaging: a. For large-area scans (millimeter-scale) to locate regions of interest, use a standard silicon nitride probe (e.g., SCANASYST-AIR) in tapping mode. b. For high-resolution imaging of cellular arrangements and flagella, switch to a sharp, high-resonant-frequency probe (e.g., RTESPA-300). Use tapping mode in liquid to minimize forces [12].
- Nanomechanical Mapping: a. Switch to a colloidal probe (e.g., CP-PNPL-BSG) for quantitative force mapping. b. Perform force-volume mapping or a high-speed spectral mode across the area of interest, ensuring indentation is limited to 10-15% of the sample thickness to minimize substrate effects [15] [13].
- Data Analysis: Use machine learning-based segmentation or standard analysis software (e.g., AFMech Suite) to stitch large-area images, detect cells, classify features, and extract mechanical properties from force curves [12] [15].
Table 2: Research Reagent Solutions - Essential AFM Probes for Biofilm Research
| Probe Type / Model | Key Specifications | Primary Function in Biofilm Research |
|---|---|---|
| Sharp Tapping Mode Probe | High resonant frequency (>300 kHz), low force constant (~5 N/m), tip radius <10 nm | High-resolution imaging of bacterial cell walls, flagella, pili, and fine EPS fibers [12] [14]. |
| Colloidal Probe | Spherical tip (radius 0.5-5 µm), moderate force constant (~0.1 N/m) | Quantitative nanomechanical mapping (modulus, adhesion); minimizes indentation damage and simplifies contact mechanics on soft, adhesive EPS [15] [13]. |
| High-Aspect-Ratio Probe | Tip height >10 µm, tip radius <10 nm | Probing deep into pores, channels, and crevices within the biofilm 3D structure without artifact generation [14]. |
| Soft Static Lever | Very low force constant (0.01 - 0.5 N/m) | High-sensitivity force spectroscopy on extremely soft regions to measure weak adhesion forces and map ultralow elastic moduli [13]. |
Probe Selection Workflow Diagram
The following diagram visualizes the decision-making process for selecting the optimal AFM probe based on your experimental goal.
Atomic Force Microscopy (AFM) is a powerful tool for studying the intricate architecture and mechanical properties of live biofilms at the nanoscale. However, the heterogeneous and viscoelastic nature of biofilms presents unique challenges, often leading to imaging artifacts that can distort biological interpretation. This technical support guide provides a focused comparison of the primary AFM imaging modes—Contact, Tapping, and PeakForce Tapping—for researchers aiming to minimize artifacts and obtain reliable data from live biofilm experiments.
AFM Imaging Modes: A Comparative Guide
The choice of AFM imaging mode is critical for successful live biofilm analysis. The table below summarizes the key characteristics, advantages, and limitations of each mode.
Table 1: Comparison of AFM Imaging Modes for Live Biofilms
| Feature | Contact Mode | Tapping Mode | PeakForce Tapping Mode |
|---|---|---|---|
| Basic Principle | Tip is in constant contact with the sample surface [16]. | Cantilever oscillates, tip intermittently contacts ("taps") the surface [16] [17]. | Controlled, periodic "tapping" where the tip engages the surface at a precise, low force [18]. |
| Tip-Sample Interaction | High; constant physical contact generates significant lateral (frictional) forces [16]. | Low; vertical oscillation minimizes lateral forces, reducing sample damage [16] [17]. | Very Low; direct control of the peak interaction force for each tap, virtually eliminating lateral forces [18]. |
| Typical Forces | 1-100 nN [16] | Lower than Contact Mode; controlled by amplitude damping. | Precisely controlled, typically at the pico- to nano-newton scale. |
| Optimal Cantilever Stiffness | Low (C ≤ 1 N/m) [16] | High (C ~ 40 N/m) [16] | Medium to High; optimized for force control. |
| Best For (Sample Type) | Hard, flat, and robust surfaces [16]. | Soft, adhesive, and loosely bound samples; excellent for heterogeneous biofilms [17]. | Very soft, delicate, and highly heterogeneous samples; ideal for live cells and hydrated biofilm matrices [18]. |
| Key Advantages | - Simple operation [16]- Enables certain electrical modes (C-AFM, TUNA) [16]- Measures lateral friction forces. | - Reduces sample damage and deformation [16] [17]- Suitable for rough and adhesive surfaces [16]- Enables Phase Imaging for material contrast [16] [17]. | - Superior force control for minimal sample disturbance [18]- Directly and quantitatively maps nanomechanical properties (e.g., adhesion, modulus) simultaneously with topography [18]. |
| Common Artifacts & Challenges | - Streaks, sample deformation, or complete removal of soft biofilm features [17]- High tip and sample wear [16]. | - Instability on very soft or adhesive regions if parameters are not optimized.- Phase images can be difficult to interpret quantitatively. | - Requires careful tuning of the peak force setpoint to balance image quality and sample protection. |
Diagram 1: A workflow for selecting the optimal AFM imaging mode for biofilm samples.
Frequently Asked Questions (FAQs) and Troubleshooting
Q1: My Tapping Mode images of a live biofilm appear blurry and unstable. The cantilever oscillation often dies out completely. What could be the cause?
- Problem: This is a classic symptom of high adhesion between the AFM tip and the hydrated, sticky EPS matrix of the biofilm. The excessive adhesive forces dampen the cantilever's oscillation amplitude beyond the feedback loop's ability to recover.
- Solution:
- Verify Cantilever: Ensure you are using a stiff cantilever (e.g., ~40 N/m) as recommended for Tapping Mode to overcome adhesion [16].
- Optimize Setpoint: Gradually increase the amplitude setpoint to reduce the tip-sample interaction time and force. Be cautious not to set it too high, which can lead to loss of contact and noisy images.
- Consider PeakForce Tapping: Switch to PeakForce Tapping Mode, which is specifically designed to handle highly adhesive samples by directly controlling and limiting the maximum force applied during each tap, preventing the tip from getting stuck [18].
Q2: I am using Contact Mode, and my scans are consistently streaking in the fast-scan direction. Furthermore, the biofilm structure appears to be "smeared." What is happening?
- Problem: This indicates excessive lateral (shear) forces are deforming or even removing the soft biofilm material during scanning. The tip is physically dragging the biofilm components across the surface.
- Solution:
- Reduce Applied Force: Immediately lower the deflection setpoint to minimize the normal force, which in turn reduces lateral forces.
- Switch to a Dynamic Mode: Contact Mode is generally unsuitable for delicate biofilms. The definitive solution is to switch to a dynamic mode like Tapping Mode or PeakForce Tapping, which minimize lateral forces by using vertical oscillations [16] [17] [18].
- Check Immobilization: Ensure your biofilm is adequately immobilized on the substrate to withstand the minimal remaining forces from a gentler mode.
Q3: How can I quantitatively map the elasticity of a live biofilm simultaneously with its topography?
- Answer: PeakForce Tapping Mode is the ideal choice for this application. In this mode, a force-distance curve is captured at every pixel of the image. The system can fit these curves to mechanical models (e.g., Hertz model) to generate quantitative maps of elastic modulus (stiffness), adhesion, and dissipation alongside the topographical image [18]. While force spectroscopy can be performed in other modes, PeakForce Tapping integrates it directly into high-resolution imaging.
Q4: My phase image on a heterogeneous biofilm shows strong contrast, but I am unsure how to interpret it. Is the contrast related to material properties?
- Answer: Yes, phase contrast in Tapping Mode is sensitive to differences in the sample's mechanical and adhesive properties. However, it is a qualitative measure. A darker phase shift often indicates a more adhesive or dissipative (softer) region, while a brighter shift can indicate a more elastic (stiffer) region [17]. For quantitative data, you must use PeakForce Tapping-based mechanical mapping. When reporting, always note that phase imaging provides qualitative material contrast, not absolute values.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 2: Key Reagent Solutions for AFM Biofilm Studies
| Item | Function in Experiment | Key Considerations |
|---|---|---|
| PFOTS-treated Glass | Creates a hydrophobic surface to promote bacterial adhesion for early-stage biofilm studies [3]. | Provides a uniform, defined surface chemistry to study attachment dynamics. |
| Polydimethylsiloxane (PDMS) Stamps | Microfabricated stamps with pores to physically trap and immobilize microbial cells for stable imaging in liquid [17]. | Crucial for preventing cells from being displaced by the scanning tip. Pore size must match the target cell diameter. |
| Poly-L-Lysine | A chemical immobilizer that creates a positively charged surface to enhance electrostatic attachment of (typically negatively charged) bacterial cells [17]. | A common and easy-to-use method, but can potentially alter surface physicochemical properties. |
| Silicon Nitride Cantilevers | The core sensing component of the AFM. Material and shape are chosen for biological compatibility and specific imaging modes [19]. | Rectangular or triangular shapes are common. Softer levers (< 1 N/m) are used for contact and force spectroscopy on soft samples, while stiffer levers (~40 N/m) are for Tapping Mode [16] [19]. |
| Spherical Tip Probes | Cantilevers with a microsphere attached to the end, used for force spectroscopy and nanoindentation [19]. | The defined geometry simplifies contact mechanics modeling for quantitative measurement of biofilm adhesion and viscoelasticity [20]. |
| Physiological Buffer (e.g., PBS) | Maintains biofilm hydration and native state during imaging in liquid. | Essential for live biofilm studies to prevent dehydration and preserve physiological function. |
Diagram 2: Key experimental workflow for AFM analysis of live biofilms, from preparation to analysis.
Leveraging Large-Area Automated AFM and Machine Learning for Seamless Stitching and Analysis
Frequently Asked Questions (FAQs) and Troubleshooting
Q1: My large-area stitched AFM image shows repetitive, unnatural patterns. What could be the cause? This is typically a tip artifact, often caused by a contaminated or broken AFM probe. A blunt or dirty tip can cause structures to appear larger than they are and fine details to be duplicated across the image [4].
- Solution:
- Replace the AFM probe: Use a new, sharp probe to see if the issue disappears [4].
- Regular inspection: Implement a protocol to regularly inspect and clean probes, especially when working with heterogeneous biofilm samples that can leave residue.
Q2: I am having difficulty accurately imaging the deep, porous structures of my biofilm. The trenches appear shallow and poorly resolved. This problem usually stems from using an AFM probe with an inappropriate shape or low aspect ratio. Standard pyramidal tips cannot reach the bottom of deep, narrow features [4].
- Solution:
- Switch to a high-aspect-ratio (HAR) conical probe: Conical tips are superior for resolving steep-edged features and deep trenches common in biofilm architecture [4].
Q3: After an automated tip approach, my image is blurry and lacks nanoscale detail, as if the tip isn't in proper contact. This indicates false feedback, where the system mistakenly believes the tip is in contact. Common causes are a thick surface contamination layer or electrostatic forces between the cantilever and sample [21].
- Solution:
- Adjust the setpoint: Increase the tip-sample interaction force. In vibrating (tapping) mode, decrease the setpoint value; in non-vibrating (contact) mode, increase it [21].
- Reduce electrostatic forces: Create a conductive path between the cantilever and sample, or use a stiffer cantilever to minimize the effect of surface charge [21].
- Improve sample preparation: Ensure protocols minimize loosely adhered material and contamination [4].
Q4: My large-area scan shows repetitive lines across the image, distorting the data. This is often due to electrical noise or laser interference [4].
- Solution:
- Identify the source: Check if the noise frequency is 50 Hz (or 60 Hz), which would indicate electrical noise from building circuits [4].
- Use reflective coatings: Employ probes with a reflective coating (e.g., gold or aluminium) to prevent laser light reflecting off the sample from interfering with the signal [4].
- Change scanning times: Image during quieter periods (e.g., early mornings) when electrical noise may be lower [4].
Q5: How can I ensure my machine learning model accurately identifies and classifies cells in a large-area AFM scan? Accuracy depends on high-quality training data and a robust model.
- Solution:
- Leverage transfer learning: Adapt a pre-trained model (like YOLOv3) for cell detection to low-quality images from an AFM stage camera, which requires limited new training data [22].
- Implement a closed-loop control: Use the ML model's output to directly control the AFM scanner trajectory, creating a feedback loop that ensures the probe navigates to the correct locations for verification and measurement [22].
Troubleshooting Guide for Common AFM Artifacts
The table below summarizes common issues, their causes, and solutions specifically for heterogeneous biofilm research.
| Problem | Cause | Solution |
|---|---|---|
| Tip Artifacts (e.g., duplicated features) [4] | Contaminated or broken AFM probe. | Replace with a new, sharp probe. |
| Poor Trench Resolution [4] | Low-aspect-ratio or pyramidal tip geometry. | Use a High-Aspect-Ratio (HAR) conical tip. |
| False Feedback (blurry images) [21] | Tip trapped in contamination layer or electrostatic forces. | Adjust setpoint; use stiffer lever; create conductive path. |
| Streaks & Blurred Lines [4] | Environmental vibrations or loose sample contamination. | Use anti-vibration table; ensure sample is securely prepared. |
| Repetitive Lines (Noise) [4] | Electrical noise (50/60 Hz) or laser interference. | Image during low-noise periods; use probes with reflective coating. |
Experimental Protocol: Automated Large-Area AFM for Biofilm Analysis
This protocol is adapted from the study on Pantoea sp. YR343 biofilm assembly [3].
1. Objective To capture high-resolution, millimeter-scale topographical images of early-stage biofilm formation, enabling the analysis of spatial heterogeneity, cellular orientation, and the role of appendages like flagella.
2. Materials and Reagents
- Bacterial Strain: Pantoea sp. YR343 (gram-negative, rod-shaped, motile with peritrichous flagella) and a flagella-deficient control strain [3].
- Growth Medium: Appropriate liquid growth medium [3].
- Substrate: PFOTS-treated glass coverslips or silicon substrates [3].
- Imaging Instrument: Atomic Force Microscope equipped with a large-area automated scanning stage.
3. Methodology
- Sample Preparation:
- Place PFOTS-treated glass coverslips in a petri dish.
- Inoculate the dish with Pantoea cells suspended in the liquid growth medium.
- Incubate for selected time points (e.g., 30 minutes for initial attachment; 6-8 hours for cluster formation).
- At each time point, remove a coverslip and gently rinse it with buffer to remove unattached cells.
- Air-dry the sample before AFM imaging [3].
- Automated Large-Area AFM Imaging:
- System Calibration: Ensure the AFM is properly calibrated.
- Region Selection: Define a millimeter-scale area for scanning.
- Automated Scanning: Initiate the automated routine to capture multiple high-resolution AFM images with minimal overlap across the defined area.
- Image Stitching: Use integrated machine learning algorithms to seamlessly stitch the individual images into a single, large-area map [3].
- Data Analysis:
- Cell Detection & Classification: Apply ML-based segmentation to automatically identify and classify individual cells [3].
- Morphometric Analysis: Extract quantitative parameters such as cell count, confluency, shape, and orientation from the stitched image [3].
- Appendage Mapping: Visually identify and map fine structures like flagella, noting their interactions and distribution [3].
4. Expected Results
- After ~30 minutes: Isolated rod-shaped cells (~2 µm long, ~1 µm diameter) with flagellar appendages (~20-50 nm in height) visible [3].
- After 6-8 hours: Formation of cell clusters with a distinctive honeycomb pattern and flagella bridging gaps between cells [3].
Workflow Diagram: Large-Area AFM with ML Analysis
Research Reagent Solutions
The table below details key materials used in the featured large-area AFM biofilm experiment [3].
| Item | Function in the Experiment |
|---|---|
| PFOTS-treated Glass Coverslips | Provides a modified surface to study bacterial adhesion dynamics and the effect of surface properties on biofilm assembly. |
| Silicon Substrates | Used to create gradient-structured surfaces for combinatorial studies on how surface modifications influence bacterial attachment density. |
| Pantoea sp. YR343 | A model gram-negative, rod-shaped bacterium with peritrichous flagella, used to study the genetic regulation and structural organization of early-stage biofilms. |
| Flagella-deficient Mutant Strain | Serves as a control to confirm the identity of flagellar structures and their specific role in biofilm assembly beyond initial attachment. |
Atomic Force Microscopy (AFM) offers unparalleled capability for high-resolution, nanoscale imaging of biological samples under near-physiological conditions. For the study of heterogeneous biofilms, maintaining precise environmental control, particularly of hydration, is not merely beneficial—it is critical for preserving native structures. Traditional electron microscopy methods involve harsh chemical fixation, dehydration, and metal coating that irreversibly alter biofilm architecture [23]. In contrast, AFM enables researchers to image fully hydrated specimens, revealing authentic structural details of extracellular polymeric substances (EPS), cellular morphologies, and intricate appendages like flagella that would otherwise be collapsed or distorted [3].
The challenge for researchers lies in overcoming the technical artifacts that arise when imaging these soft, dynamic, and heterogeneous materials in fluid. This technical support center provides targeted troubleshooting guidance and FAQs to help researchers identify, understand, and correct common artifacts specific to biofilm imaging in hydrated environments, enabling more reliable data collection and interpretation.
Troubleshooting Guide: Common AFM Artifacts in Hydrated Biofilm Imaging
Problem: Excessive Noise and Unstable Imaging in Fluid
- Observed Symptoms: Blurry images, high-frequency oscillations in trace/retrace mismatch, inability to maintain stable tip-sample interaction.
- Primary Causes:
- Environmental vibrations affecting the AFM system.
- Acoustic noise from the surroundings.
- Fluid cell turbulence or bubbles in the system.
- Brownian motion of loose material or the tip in liquid [24].
- Solutions:
- Verify Anti-Vibration Setup: Ensure the anti-vibration table is functioning correctly (e.g., check gas supply if applicable) [4].
- Minimize Acoustic Noise: Use an acoustic enclosure if available. Image during quieter times (e.g., early mornings) and use "STOP AFM in progress" signs to alert colleagues [4].
- Purge Fluid Cell: Carefully degas buffers before use and ensure the fluid cell is properly sealed and purged of all air bubbles.
- Optimize Imaging Parameters: Reduce scan size and speed to improve stability in challenging conditions. For highly mobile samples, a slight chemical fixation (e.g., 0.5% glutaraldehyde) may be necessary to stabilize structures for longer observations without complete denaturation [24].
Problem: Sample Drift or Movement During Scanning
- Observed Symptoms: Elongated or smeared features, images that shift between scan lines, inability to re-locate the same area.
- Primary Causes:
- Poor sample adhesion to the substrate.
- Insufficient fixation of the biofilm.
- Loose particles being pushed by the tip.
- Thermal drift from temperature fluctuations.
- Solutions:
- Optimize Substrate and Adhesion: Use freshly cleaved mica or treated glass surfaces (e.g., PFOTS-treated) to enhance cell attachment [3]. For powders or loose cells, resuspend in a clean solvent and deposit onto the substrate, followed by thorough drying to fix particles before hydrating for imaging [25].
- Secure Sample Mounting: Use a suitable adhesive to firmly secure the sample to the mounting stub.
- Allow Thermal Equilibrium: After introducing liquid into the cell, allow the system to equilibrate for at least 20-30 minutes to minimize thermal drift.
- Use Appropriate Imaging Mode: Switch from contact mode to a dynamic mode like TappingMode or PeakForce Tapping to minimize lateral forces that can displace loosely bound material [26] [4].
Problem: Loss of Resolution and Failure to Resolve Fine Structures
- Observed Symptoms: Inability to visualize expected fine details like flagella, EPS strands, or membrane proteins; images appear "soft" or blurred.
- Primary Causes:
- Blunt or contaminated AFM probe.
- Excessive imaging force compressing soft biofilm features.
- Biofilm surface too soft or viscoelastic.
- Insufficient resolution of the chosen probe.
- Solutions:
- Probe Selection: Use sharp, high-resolution probes designed for biological imaging. Conical tips are often superior to pyramidal ones for resolving fine features [4].
- Optimize Imaging Force: Carefully adjust the setpoint (in TappingMode) or PeakForce Setpoint to use the minimum possible force. A slight increase in force may sometimes clear away obscuring material to reveal underlying structures, as seen with hydrated collagen fibrils [24], but excessive force will damage the sample.
- Verify Probe Condition: Image a known, sharp test sample (e.g., a grating) to check for tip degradation or contamination. Replace the probe if artifacts are found [4].
Frequently Asked Questions (FAQs)
Q1: Why is it so difficult to image my unfixed, fully hydrated biofilm samples?
Hydrated biofilms are inherently soft, dynamic, and sensitive to the AFM tip's interaction forces. Unfixed specimens can begin to dissociate in buffer, becoming unstable within minutes [24]. Furthermore, turbulence and Brownian motion in liquid create significant noise [24]. A compromise is often necessary: very mild fixation (e.g., low-concentration glutaraldehyde) can stabilize the structure for the duration of the scan while preserving much of the native architecture.
Q2: What is the best AFM mode for imaging delicate biofilms in fluid without damaging them?
TappingMode (a dynamic, intermittent contact mode) and PeakForce Tapping are highly recommended. TappingMode significantly reduces lateral forces compared to contact mode, preventing sample damage and displacement [26]. PeakForce Tapping goes a step further by directly controlling and minimizing the maximum force applied to the sample at every pixel, enabling imaging with forces as low as ~10 pN, which is ideal for pristine imaging of soft biological materials [26].
Q3: My biofilm is heterogeneous over large areas, but AFM only scans small regions. How can I link cellular-scale details to the larger community structure?
This is a recognized limitation of conventional AFM. To address this, researchers are now developing automated large-area AFM approaches. These methods stitch together hundreds of high-resolution images over millimeter-scale areas, aided by machine learning for seamless stitching and analysis. This provides a detailed view of spatial heterogeneity and organization previously obscured by the small scan sizes of traditional AFM [3].
Q4: How can I be sure that the filamentous structures I'm seeing are bacterial flagella and not imaging artifacts?
Artifact identification is crucial. To confirm structures like flagella:
- Correlate with genetics: Image a flagella-deficient mutant strain under identical conditions. The absence of the filamentous structures in the mutant strongly confirms their identity as flagella [3].
- Check dimensions: Measure the height (often ~20-50 nm) and length of the appendages. True flagella will have consistent dimensions and often originate from a cell pole or are peritrichous [3].
- Assess reproducibility: Ensure the structures are visible in multiple scans, on different cells, and with different probes to rule out a tip artifact.
Experimental Protocol: Imaging Hydrated Biofilm Ultrastructure
This protocol outlines the key steps for preparing and imaging biofilm samples in fluid to preserve native ultrastructure, based on methodologies adapted from successful AFM studies of hydrated biological specimens [3] [23] [24].
Sample Preparation Workflow
Critical Parameters for Hydrated Imaging
Table 1: Key Parameters for AFM Imaging of Hydrated Biofilms
| Parameter | Recommended Setting | Rationale |
|---|---|---|
| Imaging Mode | TappingMode or PeakForce Tapping | Minimizes lateral and normal forces, preventing sample damage and displacement [26]. |
| Scan Size | Start small (1-5 µm) before scaling up | Ensures stability and allows for optimization of parameters on a manageable area. |
| Scan Rate | 0.5 - 1.5 Hz | Balances data acquisition speed with sufficient tracking of surface topography. |
| Setpoint/Peak Force | As low as possible while maintaining engagement | Preserves soft, native structures by minimizing applied force. |
| Buffer | Appropriate physiological buffer (e.g., PBS) | Maintains biofilm viability and native structure. Always degas before use. |
| Cantilever Spring Constant | 0.1 - 0.7 N/m (soft levers) | Suitable for interacting with soft biological samples without excessive indentation. |
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 2: Key Research Reagents and Materials for Hydrated Biofilm AFM
| Item | Function & Importance | Specific Examples / Notes |
|---|---|---|
| Freshly Cleaved Mica | An atomically flat, negatively charged substrate ideal for adsorbing cells and biomolecules. | Provides a consistent, clean surface for initial attachment studies [25]. |
| Chemically Treated Coverslips | Modified surfaces to study specific biofilm-surface interactions. | PFOTS-treated glass used to study Pantoea sp. YR343 attachment and patterning [3]. |
| High-Resolution Probes | Conical or sharpened silicon nitride probes for resolving fine details. | Essential for visualizing flagella (~20-50 nm height) and other nanoscale features [3] [4]. |
| Physiological Buffers | To maintain native conditions and hydration during imaging. | PBS, Tris, or HEPES buffers; must be degassed to prevent bubble formation in the fluid cell. |
| Mild Fixatives | To stabilize ultra-soft structures for duration of scan without full denaturation. | Low-concentration glutaraldehyde (0.5%) used to stabilize hydrated rat tail tendon [24]. |
| Optimal Cutting Temperature (OCT) Compound | For cryo-preservation and cryo-sectioning of tissue-supported biofilms. | Preserves native biomolecular structures in tissue sections for subsequent AFM analysis [23]. |
Protocol for Cohesive Energy and Nanomechanical Mapping to Ensure Quantitative Accuracy
Atomic Force Microscopy (AFM) provides powerful capabilities for quantifying the nanomechanical properties and cohesive strength of biofilms, which are critical for understanding their resilience and detachment behavior. However, achieving quantitative accuracy is challenging due to the inherent heterogeneity of biofilms, the complexity of tip-sample interactions, and various sources of artifacts. This guide provides standardized protocols and troubleshooting for accurate measurement of biofilm cohesive energy and nanomechanical properties, focusing on the PeakForce QNM (Quantitative Nanomechanical Mapping) mode and related techniques.
Table: Key AFM Modes for Biofilm Property Quantification
| AFM Mode | Measured Properties | Primary Application | Throughput |
|---|---|---|---|
| PeakForce QNM | Elastic Modulus, Adhesion, Dissipation, Deformation [27] | High-resolution nanomechanical mapping | Medium-High [27] |
| Force Volume | Force-distance curves at each pixel [27] | Nanomechanical mapping | Low (Slow) [27] |
| Photothermal OFF-Resonance Tapping (PORT) | Nanomechanical properties [28] | High-speed nanomechanical mapping | High [28] |
| Contact Mode Friction | Frictional energy dissipation [29] | Biofilm cohesive energy measurement | Medium |
Experimental Protocols
Protocol 1: Measuring Biofilm Cohesive Energy via AFM Abrasion
This protocol, adapted from Ahimou et al. (2007), measures the cohesive energy of a moist biofilm by quantifying the volume removed and the frictional energy dissipated during controlled scanning [29].
Materials & Reagents:
- Biofilm Sample: Grown on a suitable substrate (e.g., polyolefin membrane) [29].
- AFM with Humidity Control: Chamber maintained at ~90% relative humidity to preserve biofilm hydration [29].
- AFM Probe: V-shaped Si3N4 cantilevers with pyramidal, oxide-sharpened tips (e.g., model NPS). Spring constant must be known (e.g., 0.58 N/m) [29].
Procedure:
- Sample Equilibration: Equilibrate the moist biofilm sample in a chamber with saturated NaCl solution (~90% humidity) for 1 hour prior to measurement [29].
- Initial Topography Imaging: Image a 5x5 μm biofilm region in contact mode under a minimal applied load (~0 nN) to establish a baseline topography without perturbation [29].
- Abrasion Scanning: Zoom into a 2.5x2.5 μm sub-region within the previously scanned area. Perform repeated raster scans (e.g., 4 scans) at an elevated load (e.g., 40 nN) and a scan velocity of 50-100 μm/s to abrade the biofilm [29].
- Post-Abrasion Imaging: Reduce the applied load back to ~0 nN and capture a non-perturbative 5x5 μm image of the abraded region [29].
- Data Analysis:
- Volume Calculation: Subtract the post-abrasion height image from the pre-abrasion image to determine the volume of displaced biofilm [29].
- Cohesive Energy Calculation: Calculate the frictional energy dissipated during abrasion from the friction force data. The cohesive energy (η) is then computed as the ratio of frictional energy dissipated to the volume of biofilm removed, with typical values ranging from 0.10 to 2.05 nJ/μm³ for a 1-day biofilm [29].
Protocol 2: Quantitative Nanomechanical Mapping (QNM) of Biofilms using PeakForce Tapping
This protocol details the use of PeakForce Tapping to simultaneously map topography and quantitative mechanical properties of biofilms in a non-destructive manner [27].
Materials & Reagents:
- Biofilm Sample: Hydrated, grown on a rigid, flat substrate.
- AFM with PeakForce Tapping Mode: Capable of performing force curves at each pixel.
- Calibrated AFM Probe: Pre-calibrated probes are recommended for best accuracy. The choice of spring constant and tip radius is critical and depends on the expected modulus of the biofilm [27].
Procedure:
- Probe and System Calibration: Accurately calibrate the cantilever's spring constant, optical lever sensitivity, and tip radius. Using pre-calibrated probes with a QR code simplifies this step and enhances accuracy [27].
- Engage and Setup: Engage the probe over a representative area of the biofilm. Set the PeakForce Tapping frequency to 1-2 kHz and adjust the peak force setpoint to the lowest possible value that maintains stable feedback. This minimizes sample deformation and potential damage [27].
- Simultaneous Topography and Force Curve Mapping: Initiate the scan. The system will perform a force curve at every pixel in the image, controlling the maximum force (peak force) applied. The feedback loop maintains this peak force constant by adjusting the tip-sample distance [27].
- On-the-Fly Data Analysis: The system analyzes each force curve in real-time to extract quantitative mechanical properties. The following data channels are typically collected simultaneously [27]:
- Topography
- Elastic Modulus (DMT Model): Fit from the retraction part of the force curve.
- Adhesion Force: Minimum force on retraction (point D).
- Dissipation: Energy dissipated per cycle, calculated from the area between the approach and retract curves.
- Deformation: Maximum sample indentation.
Table: Key Parameters for Quantitative Nanomechanical Mapping of Biofilms
| Parameter | Description | Impact on Measurement | Optimization Tip |
|---|---|---|---|
| Peak Force Setpoint | Maximum force applied during each tap [27]. | High force causes damage and over-deformation; low force loses feedback. | Use the lowest stable setpoint. |
| Tip Geometry & Spring Constant | Radius of the tip and stiffness of the lever. | A blunt tip or stiff lever reduces spatial resolution and can damage soft samples. | Use sharp, soft levers (k ~0.1-1 N/m) for soft biofilms [27]. |
| Scan Rate | Speed of tip raster motion. | Too fast leads to poor force curve sampling and inaccurate feedback. | Start low (e.g., 0.5-1 Hz); increase if data quality allows. |
| Force Curve Analysis Model | Model used to fit modulus (e.g., DMT, Hertz). | An incorrect model leads to inaccurate modulus values. | Use the DMT model for biofilms, which accounts for adhesion. |
Troubleshooting Common Artifacts and Ensuring Quantitative Accuracy
Topography and Structural Artifacts
Problem: "My image has streaks/smearing, and biofilm features appear distorted."
- Cause: High lateral forces during scanning, especially in contact mode, can drag soft biofilm components [26].
- Solution: Switch from contact mode to a dynamic mode like TappingMode or PeakForce Tapping. These modes eliminate lateral forces by intermittently contacting the sample, preserving fragile biofilm structures [26] [27].
Problem: "The biofilm appears flattened, and fine EPS structure is not resolved."
- Cause: The peak force setpoint is too high, causing excessive indentation and sample deformation. A dull tip can also increase contact area, reducing resolution [27].
- Solution: Progressively lower the peak force setpoint until feedback becomes unstable, then increase slightly. Use sharp, new tips for high-resolution imaging.
Problem: "Large-area images are stitched incorrectly, showing discontinuities."
- Cause: Traditional AFM has a small scan range, making large-area imaging difficult. Manual stitching of multiple images can lead to errors [3] [30].
- Solution: Use an automated large-area AFM platform. Integrate machine learning algorithms for seamless image stitching and cell detection, which can accurately assemble high-resolution maps over millimeter-scale areas [3] [30].
Nanomechanical and Cohesive Property Artifacts
Problem: "Measured elastic modulus values are inconsistent or unrealistically high for a soft biofilm."
- Cause 1: Tip contamination (e.g., by EPS) changes the tip geometry and contact mechanics.
- Solution: Clean the tip with solvents (e.g., ethanol) or use UV-ozone cleaning. Regularly verify tip shape.
- Cause 2: Inaccurate probe calibration, especially of the tip radius and spring constant.
- Solution: Use pre-calibrated probes. Re-calibrate the spring constant if results are suspicious. The accuracy of quantitative techniques depends on the accuracy of all other variables in the system [27].
- Cause 3: Hydrodynamic squeezing effects in liquid, which can artificially increase the measured force.
- Solution: Use low approach/retract velocities or sinusoidal excitation (as in WaveMode/PORT) to minimize viscous forces [28].
- Cause 1: Tip contamination (e.g., by EPS) changes the tip geometry and contact mechanics.
Problem: "Cohesive energy measurements show high variability between locations on the same biofilm."
- Cause: Biofilms are inherently heterogeneous. A single measurement is not representative [3] [29].
- Solution: Increase sample size (n). Perform cohesive abrasion tests or nanomechanical maps at multiple, randomly selected locations across the biofilm to capture the true spatial heterogeneity and obtain statistically robust average values [3] [29].
Frequently Asked Questions (FAQs)
Q1: How long does a typical AFM experiment for nanomechanical mapping of a biofilm take?
- A: A single high-resolution force map using PeakForce QNM can take several minutes. A full experiment, including sample preparation, calibration, and multiple maps to ensure statistical significance, can take a few hours. High-speed methods like Photothermal PORT can reduce this time by an order of magnitude [28] [25].
Q2: Can these measurements be performed under physiological (liquid) conditions?
- A: Yes. This is a key advantage of AFM. Both PeakForce QNM and Photothermal PORT can be operated in liquid, allowing for the characterization of biofilms in their native, hydrated state [28] [27].
Q3: My biofilm is very heterogeneous. How can I get a representative measurement?
- A: Use large-area automated AFM to survey big areas (millimeter-scale) and identify different regions of interest [3] [30]. Then, employ machine learning-based segmentation and classification to automatically analyze thousands of individual cells and features, providing statistically powerful property distributions across the entire community [3] [31].
Q4: What is the difference between adhesion force and cohesive energy?
- A: Adhesion force (measured in nN) is the pull-off force between the AFM tip and a specific point on the sample surface [27]. Cohesive energy (measured in nJ/μm³) is an intrinsic material property representing the energy required to break internal bonds and separate a unit volume of the biofilm material, derived from friction and wear measurements [29].
The Scientist's Toolkit: Essential Research Reagents & Materials
Table: Essential Materials for AFM Analysis of Biofilm Cohesion and Mechanics
| Item | Function/Description | Example/Specification |
|---|---|---|
| Pre-Calibrated AFM Probes | Cantilevers with pre-determined spring constant and tip radius for quantitative accuracy [27]. | Bruker ScanAsyst-Air (for air), MSNL (for high resolution), MLCT (for liquid). |
| Chemically Functionalized Tips | Probes with specific chemistry to probe ligand-receptor interactions within the EPS [29]. | Colloidal tips with coated functional groups (e.g., -COOH, -NH2). |
| Rigid & Flat Substrates | Supports for biofilm growth that minimize substrate compliance effects on mechanical measurements. | Freshly cleaved mica, glass, silicon wafers, PFOTS-treated coverslips [3]. |
| Humidity Control System | Prevents dehydration of moist biofilms during ex-situ measurements, preserving native properties [29]. | AFM chamber with humidity controller (e.g., 90% RH). |
| Calibration Samples | Reference materials with known mechanical properties to verify AFM performance and probe calibration. | Polystyrene, Polyethylene, PDMS. |
Troubleshooting Common AFM Biofilm Artifacts: A Step-by-Step Optimization Guide
Diagnosing and Correcting Tip Convolution Artifacts on Flagella and EPS Fibrils
Atomic Force Microscopy (AFM) has become an indispensable tool in biofilm research, enabling scientists to resolve the complex architecture of microbial communities at the nanoscale. Its capability to operate under physiological conditions provides unparalleled insights into the structural and functional properties of biofilms at the cellular and sub-cellular level [3]. Specifically, AFM allows for the visualization of critical biofilm components such as flagella—thin filamentous appendages critical for bacterial motility and surface attachment—and extracellular polymeric substance (EPS) fibrils that form the scaffold of the biofilm matrix [3]. These structures are essential for understanding biofilm assembly, resilience, and function.
However, a fundamental challenge persists: tip convolution artifacts. These artifacts arise because every AFM image is a convolution of the ideal sample topography with the finite geometry of the scanning probe tip [8] [32]. This effect is not merely a minor inconvenience; it represents a significant source of measurement error that can distort the apparent size, shape, and even the very presence of nanoscopic features. When imaging high-aspect-ratio structures like flagella (typically 20–50 nm in height) and delicate EPS fibrils, the tip geometry can dramatically alter their apparent dimensions, leading to incorrect biological interpretations [3] [33]. For researchers investigating heterogeneous biofilm samples, diagnosing and correcting these artifacts is not an optional step but a necessary prerequisite for deriving accurate, quantitative data from AFM images.
Understanding and Diagnosing Tip Convolution Artifacts
Tip convolution occurs when the physical dimensions of the AFM tip are comparable to or larger than the features being imaged. Instead of tracing the true surface profile, the tip apex, or worse, its sidewalls, make contact with the sample, resulting in a topography image that represents the shape of the tip as much as the shape of the sample [8] [32]. The severity of this effect is influenced by the tip's geometry, including its apex radius, aspect ratio, and sidewall angle [4] [33].
Common Artifacts on Heterogeneous Biofilm Features
The heterogeneous nature of biofilms, which combine cellular structures, flagella, and a complex EPS matrix, makes them particularly susceptible to a range of convolution artifacts. The table below summarizes the key artifacts, their causes, and how to identify them.
Table 1: Common AFM Tip Convolution Artifacts and Their Identification in Biofilm Samples
| Artifact Manifestation | Primary Cause | Effect on Biofilm Features | Diagnostic Clues |
|---|---|---|---|
| Apparent Widening of Nanofilaments | Blunt tip (large apex radius) or low-aspect-ratio tip [4] [34]. | Flagella and EPS fibrils appear significantly wider than their true diameter [3]. | Measured widths of fibrils are uniform and match the tip's apex diameter. |
| Loss of Structural Resolution | Tip geometry preventing access into narrow gaps [33]. | Fine details of the honeycomb pattern in bacterial clusters are obscured [3]. | Merged topography where distinct features appear connected. |
| "Tail" or "Shadow" Artifacts | Steep sidewalls of the tip contacting sample protrusions [33]. | Elongated streaks or shadows appear adjacent to bacterial cells. | Asymmetric, repeating patterns in the fast-scan direction. |
| Inaccurate Depth Measurement | Tip unable to reach the bottom of narrow trenches [4]. | The depth of pores in the EPS matrix is underestimated. | Trench depths are shallower than expected, with rounded bottoms. |
A Systematic Workflow for Diagnosis
Diagnosing tip convolution requires a systematic approach to distinguish genuine sample topography from artifact. The following diagram outlines a recommended diagnostic workflow.
Correction Strategies and Experimental Protocols
Once diagnosed, a combination of experimental best practices and computational correction methods can mitigate the impact of tip convolution.
Probe Selection: The First Line of Defense
The most effective way to minimize artifacts is to use a tip with an appropriate geometry. For the fine, filamentous structures found in biofilms, high-aspect-ratio (HAR) tips are essential [4]. Conical tips are often superior to pyramidal ones for resolving high features, and tetrahedral tips with steep edges have been shown to produce artifact-free topography on dense arrays of nano-features [4] [33]. The development of specialized tips like carbon nanotube (CNT) probes can further improve resolution, though their cost can be prohibitive [32].
Computational Surface Reconstruction
When the raw AFM data is affected by convolution, computational methods can reconstruct a more accurate surface profile. These algorithms are based on the principle of mathematical erosion, effectively deconvoluting the tip shape from the image [8].
Protocol: Surface Reconstruction using Gwyddion
Determine Tip Geometry: The first step is to define the tip's shape.
- Method A (Blind Estimation): In Gwyddion, use
Tools → Tip Shape → Blind Estimation. This algorithm iteratively analyzes the image data itself to estimate the tip's structure. It is recommended to run a "Partial estimation" first, followed by a "Full estimation" for a more refined result [8]. - Method B (Model Input): If the manufacturer's specifications are known, use
Tools → Tip Surface → Model Tip. For a standard conical tip, input the tip slope and the apex radius [8].
- Method A (Blind Estimation): In Gwyddion, use
Perform Surface Reconstruction: With the tip shape defined, proceed to
Tools → Tip Shape → Surface Reconstruction. This function applies an erosion algorithm to remove the tip's contribution from the image, revealing a closer approximation of the true sample topography [8].Generate a Certainty Map: To identify regions of the image that are irreversibly corrupted (e.g., deep pores the tip could not enter), use
Tools → Tip Shape → Certainty Map. This highlights data points where the tip did not touch the surface in a single point, indicating a total loss of information [8].
Quantitative Correction Algorithm
For simple, well-defined nanostructures, a geometrical correction can be applied. The relationship between the real and measured dimensions can be described mathematically. For a spherical tip apex scanning a cylindrical structure like a flagellum, the measured width ((Wm)) is related to the real width ((Wr)) and the tip radius ((R_t)) by:
(Wm \approx Wr + 2R_t)
This simple formula demonstrates that the broadening effect is directly proportional to the tip size, allowing for a straightforward numerical correction if the tip radius is known [35].
FAQs: Troubleshooting Common Problems
Q1: My AFM images of bacterial flagella show uniform widths that look too large. What is the most likely cause? This is a classic sign of tip convolution caused by a blunt or contaminated tip. The measured width of the flagella is dominated by the geometry of the tip apex rather than the actual sample. The solution is to replace the probe with a new, sharp one and, if possible, use a tip with a high aspect ratio [4] [34].
Q2: How can I verify the quality and sharpness of my AFM probe before imaging? The most reliable method is to image a reference sample with sharp, well-defined features of known dimensions, such as a test grating with steep edges. The resulting image will clearly reveal the shape and effective sharpness of the tip. If such a sample is unavailable, the blind tip estimation algorithm in software like Gwyddion can provide an estimate based on any suitable image data [8] [32].
Q3: When imaging soft, hydrated EPS in liquid, I get streaks and unstable imaging. Is this a tip artifact? While this can be related to tip contamination, streaks are more commonly caused by environmental noise/vibration or the tip moving loose surface material. Ensure the AFM is on a stable, active vibration isolation table. For biological samples in liquid, employing a gentle tapping mode or fast force-distance curve-based imaging (like PeakForce Tapping) can drastically reduce lateral forces and minimize sample disturbance [4] [36].
Q4: Can I completely eliminate tip convolution effects? It is impossible to eliminate the effect entirely, as the tip and sample must always interact. However, its impact can be minimized to a negligible level by combining optimal probe selection, careful experimental setup, and post-processing correction. The goal is to manage and correct for the artifact to extract accurate quantitative data [8] [32].
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for High-Resolution AFM of Biofilms
| Item | Function/Benefit | Application Notes |
|---|---|---|
| High-Aspect-Ratio (HAR) Conical Tips | Reduces obstructive effects from tip sidewalls, allowing more accurate profiling of tall, narrow features like flagella [4] [33]. | Superior to pyramidal tips for probing complex biofilm topography. |
| Tetrahedral Tips | Steep edges minimize artifacts when imaging dense arrays of features, providing more reliable topography [33]. | Ideal for mapping the organized honeycomb patterns of bacterial clusters. |
| Reference Sample (e.g., Sharp Grating) | Essential for independent verification of tip sharpness and geometry, and for validating correction algorithms [32]. | Use before critical experiments to confirm probe condition. |
| Software with Deconvolution Tools (e.g., Gwyddion) | Provides algorithms for blind tip estimation and surface reconstruction to correct acquired images [8]. | Open-source software like Gwyddion makes these methods widely accessible. |
| PFOTS-treated Glass Substrates | Creates a hydrophobic surface that promotes specific bacterial adhesion, facilitating the study of early attachment stages [3]. | Used in the study of Pantoea sp. YR343 biofilm assembly. |
For researchers investigating the nanoscale world of biofilms, tip convolution is a central and inescapable consideration. The artifacts it introduces can compromise the integrity of morphological data on crucial structures like flagella and EPS fibrils. A rigorous approach, combining informed probe selection, optimized imaging protocols, and robust computational correction, is required to ensure data accuracy. By systematically diagnosing and correcting for these artifacts, scientists can leverage the full power of AFM to uncover reliable, quantitative insights into the structure and function of biofilms, ultimately advancing our understanding in fields ranging from environmental microbiology to antimicrobial drug development.
Minimizing Scanner Nonlinearity and Drift for Accurate Large-Area and Time-Lapse Studies
Core Concepts: Scanner Artifacts in Biofilm Research
Atomic Force Microscopy (AFM) is a powerful tool for studying the intricate architecture of biofilms at the nanoscale. However, its accuracy, especially in large-area and time-lapse studies essential for observing biofilm development, is compromised by scanner nonlinearities and drift. These artifacts can distort topographical data, leading to an inaccurate representation of the biofilm's true structure and dynamics.
- Scanner Nonlinearity primarily refers to hysteresis and creep. Hysteresis causes the scanner's position to depend on its previous movement history, leading to different forward and backward paths for the same commanded voltage. Creep is the slow, time-dependent deformation of the piezoelectric material after a sudden voltage change. In images, this manifests as distortions where the same feature appears different when scanned from left-to-right versus right-to-left [37].
- Scanner Drift describes the gradual change in the scanner's position over time, even when the commanded voltage is constant. It is often caused by temperature fluctuations, piezoelectric aging, or electronic instability. Drift is particularly detrimental to time-lapse experiments, as it causes the apparent position of features on the biofilm to shift over time, which can be mistaken for actual biological movement or growth [38].
For heterogeneous biofilms, whose functional properties are dictated by their complex spatial organization, these artifacts can lead to incorrect measurements of critical parameters such as cellular orientation, the dimensions of extracellular polymeric substance (EPS) fibers, and the evolution of pore networks, thereby undermining the validity of the research [3] [39].
Troubleshooting Guide: FAQs on Scanner Artifacts
Q1: My AFM images of bacterial biofilms show a "smearing" or "shearing" effect, where features are stretched or compressed. What is the most likely cause and how can I fix it?
A: This is a classic symptom of hysteresis in the lateral (X-Y) scanners [37].
- Quick Fix: Ensure you are using a non-resonant, force-controlled mode like PeakForce Tapping, which provides superior force control and can mitigate some image distortions. Acquire images at a moderate speed and check if the distortion changes with speed [26].
- Advanced Solution: Implement a data-driven feedforward controller. This method uses a pair of forward and backward scan lines of your biofilm sample to identify the hysteresis mapping. A genetic algorithm can solve the nonlinear optimization problem to derive parameters that pre-distort the command signals, canceling out the hysteresis effect. This sensorless method is particularly suitable for high-speed AFM on soft biological samples [37].
Q2: During long-term time-lapse imaging of biofilm formation, the field of view appears to drift, making it difficult to track the same cluster of cells. How can I stabilize the image?
A: This is caused by scanner drift, predominantly in the Z-axis but also affecting X-Y positioning [38].
- Protocol for Drift Minimization:
- Thermal Stabilization: Allow the AFM system to equilibrate in the imaging environment (e.g., on the microscope stage, within a liquid cell) for at least 1-2 hours before starting the experiment.
- Scanner Conditioning: "Wake up" the piezoelectric scanner by scanning a small area (e.g., 1x1 µm) at a relatively high speed for 10-15 minutes prior to the main experiment.
- Software Correction: Utilize the AFM's built-in drift compensation algorithms. These algorithms track specific features in successive scans and adjust the scanner position to keep them aligned.
- Hardware Consideration: For dedicated high-speed or long-term studies, invest in a scanner designed for high mechanical stability. Flexure-based, parallel-kinematic scanner designs with high resonance frequencies (>2 kHz) exhibit lower drift and are more stable at high scan speeds [38].
Q3: When I stitch multiple AFM images together to create a large-area map of a biofilm, the edges do not align perfectly. What strategies can improve stitching accuracy?
A: Misalignment is often due to a combination of nonlinearity, drift, and sample tilt.
- Strategy:
- Pre-Correct Scanner Nonlinearity: Apply the feedforward hysteresis compensation mentioned in A1 to ensure each individual tile is geometrically accurate [37].
- Maximize Overlap: Acquire adjacent images with a significant overlap (e.g., 15-25%) to provide the stitching software with ample matching features.
- Leverage Machine Learning: Employ advanced image stitching algorithms aided by machine learning. These algorithms can seamlessly fuse tiles even with minimal matching features, which is crucial for maximizing acquisition speed over large areas [3].
Experimental Protocols for Artifact Correction
Protocol 1: Data-Driven Hysteresis Identification and Feedforward Control
This protocol details the procedure for characterizing and compensating for lateral scanner hysteresis using the sample itself, without requiring external sensors [37].
- Objective: To identify the nonlinear input-output mapping of the X-scanner and generate a compensating feedforward signal.
- Materials:
- AFM system with open-access software for custom waveform generation (or a built-in feedforward control option).
- A stable, feature-rich biofilm sample or calibration grating.
- Methodology:
- Data Acquisition: Image a small, feature-rich area of your biofilm sample (or a calibration grating) at a typical scan speed and size. Acquire a complete data set including both the trace (forward) and retrace (backward) images.
- Image Analysis: Treat the forward ((U)) and backward ((V)) images as matrices. Identify matching columns (({u}{i}) and ({v}{j})) between the two images. A search algorithm finds the column in the backward image that most closely resembles each column in the forward image, creating a vector of image mappings, ({m}{tr}).
- Optimization: Formulate a nonlinear optimization problem where the parameters of a hysteresis model (e.g., a Preisach model) are adjusted until the difference between the predicted and measured mapping (({m}{tr})) is minimized. This is efficiently solved using a genetic algorithm.
- Controller Implementation: The optimized parameters define the inverse hysteresis model. This model is implemented as a feedforward controller ((K)) in series with the scanner. It pre-shapes the command signal before it is sent to the actuator, effectively linearizing its response.
The workflow below illustrates this data-driven process.
Protocol 2: Large-Area, High-Resolution Imaging with Minimal Distortion
This protocol is designed for acquiring large-area maps (up to millimeter-scale) of biofilms with nanometer resolution, which is critical for linking nanoscale cellular features to the functional macroscale organization of the film [3] [38].
- Objective: To obtain a seamless, high-resolution mosaic image over a large sample area.
- Materials:
- AFM system with a large-sample scanner and automated stage.
- Software supporting automated tile-based acquisition and ML-powered stitching.
- Methodology:
- Scanner Selection and Calibration: Use a scanner designed for large travel ranges with high resonance frequencies (e.g., >2 kHz). Calibrate the scanner using a traceable standard to ensure linearity.
- Pre-Scan Setup: Define the large area of interest on the sample. Set the overlap between adjacent tiles to ~20%. Use a non-resonant imaging mode (e.g., PeakForce Tapping) to minimize lateral forces and sample damage.
- Automated Acquisition: Run the automated large-area acquisition script. The system will sequentially image each tile.
- Image Stitching and Analysis: Use a stitching algorithm that incorporates machine learning for cell detection and classification. This helps in creating a seamless composite image and enables quantitative analysis of parameters like cell count, confluency, and orientation over the entire large area [3].
Table 1: Comparison of Scanner Performance and Correction Techniques
| Scanner Type / Technique | Max Scan Size | Key Artifact Addressed | Typical Resolution | Best Use Case in Biofilm Research |
|---|---|---|---|---|
| Standard Piezo Scanner [26] | ~100 µm | N/A | < 1 nm | High-resolution imaging of single cells or small clusters. |
| Ultra-Wide HS-AFM Scanner [38] | 36 x 36 µm² | Hysteresis, Resonances | Molecular (~4 nm) | Dynamics of large molecular assemblies and single molecules over large areas. |
| Data-Driven Feedforward Control [37] | Applicable to any size | Hysteresis | Improves effective accuracy | High-speed AFM and any study requiring high geometric fidelity in X-Y. |
| Automated Large-Area AFM [3] | Millimeter-scale | Drift, Stitching errors | Nanoscale | Mapping spatial heterogeneity across entire early-stage biofilm communities. |
Table 2: Key Research Reagent Solutions for AFM of Biofilms
| Reagent / Material | Function / Description | Application in Biofilm Research |
|---|---|---|
| Functionalized Mica (e.g., APS-mica) [40] | Provides an atomically flat, positively charged surface for sample adhesion. | Immobilization of biological samples like nucleosomes or bacterial cells for high-resolution imaging in liquid. |
| PFOTS-treated Glass [3] | A silane-based treatment that creates a hydrophobic surface. | Studying the early stages of bacterial attachment and biofilm assembly on abiotic surfaces. |
| Refolding Buffer [40] | Typically contains 2 M NaCl, Tris-HCl, EDTA; used for histone octamer assembly. | Biochemical reconstitution of protein complexes (e.g., nucleosomes) for structural dynamics studies. |
| Extracellular Polymeric Substances (EPS) [39] | Self-produced matrix of polysaccharides, proteins, and nucleic acids. | The primary target of study, as its structure and properties govern biofilm stability and mass transfer. |
The Scientist's Toolkit: Visualization of Analysis Workflow
The following workflow outlines the process of using AI-powered tools to correct artifacts and extract meaningful quantitative data from large-area AFM images of biofilms, linking raw data to biological insight.
Atomic Force Microscopy (AFM) is an indispensable tool in biofilm research, capable of revealing the intricate architecture of microbial communities at the nanoscale. However, the heterogeneous and often soft nature of biofilms presents a significant challenge. Incorrect scan parameters can lead to sample damage, distorted data, and the introduction of imaging artifacts. This guide provides targeted troubleshooting advice and FAQs to help researchers optimize key AFM parameters—setpoint, gain, and scan rate—to obtain high-fidelity images while preserving the integrity of delicate biofilm samples.
Troubleshooting Guides
Guide 1: Diagnosing and Resolving Common AFM Imaging Problems in Biofilms
Biofilm samples are prone to specific issues that can degrade image quality. The table below outlines common problems, their likely causes, and solutions.
| Problem Observed | Possible Cause | Recommended Solution |
|---|---|---|
| Blurry images, loss of detail [41] | False feedback from surface contamination or electrostatic forces [41] | Increase tip-sample interaction: decrease setpoint in TappingMode or increase it in Contact Mode [41]. |
| Streaks or horizontal lines in image [4] | Loose particles or EPS being dragged by the tip [4] | Improve sample preparation to remove loose material; ensure gentle rinsing [4]. |
| Trace and Retrace lines not aligning [42] | Scan rate too high or gains improperly set [42] | Reduce scan rate until lines overlap; then adjust Proportional and Integral gains [42]. |
| Repetitive noise patterns [4] | Electrical noise or laser interference [4] | Image during quieter times (e.g., early morning); use probes with reflective coatings [4]. |
| Sample damage, cells moved or scraped [26] | Excessive imaging force (setpoint too low in TappingMode) or high lateral forces in Contact Mode [26] | Use a gentler mode (TappingMode or PeakForce Tapping) [26]; increase setpoint to reduce force [42]. |
Guide 2: A Step-by-Step Protocol for Parameter Optimization
Follow this sequential protocol to systematically optimize your AFM parameters for stable and non-destructive imaging of biofilms [42].
Step 1: Optimize Imaging Speed / Scan Rate
- Action: Observe the Trace and Retrace height contours. If they do not closely overlap, the AFM tip is not tracking the topography correctly.
- Adjustment: Gradually reduce the Scan Rate or Tip Velocity.
- Goal: Achieve a close overlap between Trace and Retrace lines. A small offset is acceptable. Further reduction after this point unnecessarily increases acquisition time [42].
Step 2: Optimize Feedback Gains (Proportional & Integral)
- Action: Continue observing the Trace and Retrace lines.
- Adjustment: If the lines are still misaligned, gradually increase the Proportional Gain and Integral Gain.
- Goal: Bring the Trace and Retrace lines into close alignment without introducing visible noise or spikes into the image. The presence of noise indicates the gains are too high and must be reduced [42].
Step 3: Optimize the Setpoint (for TappingMode or PeakForce Tapping)
- Action: With scan rate and gains optimized, check the final tracking of the height contours.
- Adjustment: If tracking is unstable, gradually decrease the Setpoint to increase the tip-sample interaction force.
- Goal: Achieve stable tracking where Trace and Retrace lines follow each other closely. To minimize sample damage and tip wear, use the highest possible setpoint that still provides stable imaging [42].
This optimization workflow is summarized in the following diagram:
Frequently Asked Questions (FAQs)
Q1: What is the fundamental difference between Contact Mode and TappingMode, and which is better for soft biofilm samples?
A1: In Contact Mode, the probe tip is in constant physical contact with the sample, which can generate high lateral forces that distort or displace weakly attached cells and extracellular polymeric substances (EPS) [26]. TappingMode oscillates the cantilever and gently taps the surface, minimizing these lateral forces. For soft, fragile biofilms, TappingMode or the more advanced PeakForce Tapping is generally recommended to prevent sample damage [26].
Q2: My image shows repeating, unnatural shapes. What is happening?
A2: This is a classic tip artifact. It indicates that your AFM tip is either contaminated with debris from the sample or has become broken and blunt. A contaminated or broken tip will produce images that are a convolution of the tip's shape and the true sample topography. The solution is to replace the probe with a new, clean one [4].
Q3: How does "false feedback" occur, and how can I fix it?
A3: False feedback occurs when the AFM's automated tip approach is "tricked" into stopping before the probe interacts with the sample's hard surface forces. This is common in biofilms due to thick layers of soft EPS or surface contamination [41]. The probe gets trapped in this soft layer, resulting in a blurry, out-of-focus image. To fix it, you need to increase the tip-sample interaction force by decreasing the setpoint value in TappingMode [41].
Q4: What are the key considerations for preparing a biofilm sample for AFM to avoid artifacts?
A4:
- Cleanliness: Your sample must be as clean as possible. Use high-purity water (e.g., molecular biology grade) and buffers to avoid introducing particulate contaminants that can cause streaks or stick to the tip [25] [4].
- Fixation: While AFM can be performed under physiological conditions, gently fixing biofilms (e.g., with low concentrations of glutaraldehyde) can stabilize the structure and reduce the risk of disruption by the tip.
- Rinsing: Gently but thoroughly rinse the sample to remove loosely attached cells and debris that could be dragged by the tip during scanning [4].
Experimental Protocols
Protocol: Investigating Biofilm-Surface Interactions Using Large-Area AFM
1. Background and Objective: Traditional AFM is limited by a small scan range, making it difficult to link nanoscale cellular features to the macro-scale organization of a biofilm. This protocol, adapted from Millan-Solsona et al., uses an automated large-area AFM approach to study the early stages of biofilm formation across millimeter-scale areas, revealing spatial heterogeneity and organizational patterns like honeycomb structures [3] [30].
2. Materials and Reagents:
| Item | Function/Brief Explanation |
|---|---|
| PFOTS-treated glass coverslips | Creates a hydrophobic surface to promote bacterial attachment for study [3]. |
| Pantoea sp. YR343 (wild-type and flagella-deficient mutant) | Model gram-negative bacterium for studying attachment and biofilm structure. The mutant serves as a control [3]. |
| Liquid Growth Medium (e.g., BHI) | Supports bacterial growth and provides nutrients for biofilm development. |
| Atomic Force Microscope with large-area automation | Enables automated acquisition of multiple adjacent high-resolution images over a large area [3] [30]. |
| Machine Learning-based image analysis software | Stitches individual image tiles and automates cell detection, classification, and morphological analysis [3]. |
3. Methodology:
- Sample Preparation: Inoculate a petri dish containing the PFOTS-treated coverslips with Pantoea cells in liquid growth medium. At specific time points (e.g., 30 min, 6-8 h), remove a coverslip and gently rinse it with purified water or a mild buffer to remove non-adherent cells. Air-dry the sample before imaging [3].
- Large-Area AFM Imaging:
- Mount the prepared coverslip on the AFM stage.
- Define a large area (e.g., several mm²) for scanning within the software.
- Use the automated routine to acquire multiple consecutive high-resolution images with minimal overlap.
- Data Processing and Analysis:
- Image Stitching: Use computational algorithms to merge the individual image tiles into a seamless, large-area map [3] [30].
- Feature Analysis: Apply machine learning models to the stitched image to automatically identify and segment individual bacterial cells. The software can then extract quantitative data such as cell density, confluency, orientation, and the presence of appendages like flagella [3].
4. Expected Outcome: This protocol allows for the quantitative analysis of spatial heterogeneity during early biofilm formation. Researchers can identify patterns, such as the distinctive honeycomb pattern observed in Pantoea sp. YR343, and correlate nanoscale features (e.g., flagellar bridges between cells) with the larger community structure [3] [30].
The logical flow of this experimental workflow is as follows:
Strategies to Mitigate Adhesive Forces and Probe Contamination in Hydrated EPS Matrix
Technical Support Center
Frequently Asked Questions (FAQs)
FAQ 1: What are the primary causes of high adhesive forces and probe contamination when performing AFM on hydrated biofilms? High adhesive forces and probe contamination primarily result from the complex, heterogeneous nature of the extracellular polymeric substance (EPS) matrix. This gelatinous, adhesive environment causes the AFM tip to become stuck or fouled by EPS components like polysaccharides, proteins, and nucleic acids during force spectroscopy or imaging. Traditional single-cell force spectroscopy (SCFS) methods are particularly susceptible as they do not represent the realistic, community-based structure of biofilms, leading to unpredictable interactions and probe contamination [43].
FAQ 2: How can I reduce probe contamination during repeated force spectroscopy measurements on biofilm samples? Utilizing FluidFM technology is a highly effective strategy. This method uses microfluidic cantilevers where a negative pressure is applied to immobilize a probe (e.g., a single cell or a biofilm-coated bead) at the aperture. This setup creates a stable, reversible immobilization that is less damaging and contaminating than traditional chemical gluing methods. The closed fluidic system allows for the probe to be refreshed or cleaned between measurements, significantly reducing cross-contamination and buildup of EPS material on the cantilever [43].
FAQ 3: My AFM images of biofilms lack larger context. How can I correlate nanoscale features with community-scale organization? Implement a large-area automated AFM approach integrated with machine learning. This overcomes the traditional limitation of AFM's small scan range by automatically capturing and stitching together multiple high-resolution images over millimeter-scale areas. This provides a comprehensive view that links individual cellular features, like flagella, to the broader biofilm architecture, such as honeycomb patterns. Machine learning algorithms further assist in automating the analysis of these large datasets for parameters like cell count, confluency, and orientation [3] [44].
FAQ 4: How does the mechanical properties of a cell membrane influence adhesion force measurements? The cell membrane's tension and elasticity significantly impact indentation and retraction curves. Standard models like the Hertz model do not fully account for membrane properties. During retraction, bonds formed with membrane receptors can lead to the formation of long membrane tethers, manifesting as sawtooth patterns in the force-distance curve with extensions of hundreds of nanometers. Accounting for membrane tension and elasticity is crucial for accurate interpretation of single-molecule adhesion experiments [45].
Troubleshooting Guides
Problem 1: Inconsistent and Uninterpretable Sawtooth Patterns in Force-Distance Curves
- Potential Cause: The complex, stochastic disruption of bonds coupled with the extraction of the elastic cell membrane, rather than a simple reflection of bond dynamics [45].
- Solution:
- Develop a theoretical model that incorporates membrane tension, elasticity, and AFM tip geometry to simulate detachment.
- For specificity, use appropriate negative controls, such as probing a mutant cell line that lacks the receptor of interest [45].
Problem 2: Low Throughput and Representative Data from AFM Biofilm Analysis
- Potential Cause: Reliance on manual operation and small, single-point scans which cannot capture the spatial heterogeneity of a biofilm [3].
- Solution:
Problem 3: Measuring Biofilm-Cohesive Strength in Hydrated Conditions
- Potential Cause: Standard methods may require drying the sample, which alters the biofilm's native mechanical properties [29].
- Solution: Use an in-situ abrasion-based AFM method to measure cohesive energy in hydrated biofilms.
- Mount the hydrated biofilm sample in a chamber with controlled humidity (e.g., ~90%) to maintain water content.
- Collect a non-perturbative topographic image of a region at a low applied load (~0 nN).
- Switch to a smaller sub-region and abrade the biofilm under repeated raster scanning at an elevated load (e.g., 40 nN).
- Return to a low load and image the abraded region again.
- Calculate the volume of displaced biofilm from the topographic difference and the frictional energy dissipated from the friction signal. The cohesive energy (nJ/μm³) is the ratio of frictional energy to displaced volume [29].
The table below summarizes key quantitative findings from recent research on biofilm adhesion and AFM analysis.
Table 1: Quantitative Data on Biofilm Adhesion and AFM Performance
| Measurement Parameter | Reported Value / Range | Experimental Context | Source |
|---|---|---|---|
| Biofilm Cohesive Energy | 0.10 ± 0.07 nJ/μm³ to 2.05 ± 0.62 nJ/μm³ | Increases with depth in hydrated 1-day biofilms from activated sludge [29]. | |
| Bacterial Cell Dimensions (Pantoea sp. YR343) | ~2 µm length, ~1 µm diameter | Surface-attached cells visualized via high-resolution AFM [3]. | |
| Flagella Height | ~20–50 nm | Appendages observed around bacterial cells using AFM [3]. | |
| Peak Detachment Forces (CD47-SIRPα) | ~1500 pN (first peak), ~600 pN (second peak) | Force spectroscopy on red blood cells with a functionalized AFM tip [45]. | |
| Cell Surface Area | ~2 μm² | Calculated for Pantoea sp. YR343 based on AFM measurements [3]. | |
| Classification Accuracy (Biofilm Maturity) | Human: 0.77 ± 0.18ML Algorithm: 0.66 ± 0.06 | Accuracy of classifying staphylococcal biofilm images via AFM and machine learning [31]. |
Experimental Protocols
Protocol 1: FluidFM for Biofilm-Surface Adhesion Force Measurement This protocol measures adhesion forces between a biofilm and a surface, such as an anti-fouling membrane [43].
- Biofilm Probe Preparation:
- Grow bacterial biofilms on micrometer-sized, COOH-functionalized polystyrene beads for 3 hours.
- Aspirate a single biofilm-coated bead onto the aperture of a microfluidic FluidFM cantilever by applying a negative pressure via the pressure controller.
- Surface Preparation:
- Modify filtration membrane surfaces with an anti-biofouling agent (e.g., vanillin at 3 g/L in PBS).
- Force Spectroscopy:
- Approach the biofilm-attached FluidFM probe to the membrane surface in a liquid environment.
- After a set contact time, retract the probe and record the force-distance curves.
- Analyze the retraction curves for adhesion force, adhesion work, and the number of adhesion events.
- Compare results between modified and unmodified surfaces to quantify the reduction in biofouling potential.
Protocol 2: In-Situ Measurement of Biofilm Cohesive Strength This protocol details the measurement of cohesive energy within a hydrated biofilm [29].
- Sample Preparation:
- Grow a 1-day biofilm on a suitable substrate (e.g., a gas-permeable membrane in a reactor).
- Cut a hydrated sample and equilibrate it in an AFM chamber controlled at ~90% relative humidity to maintain moist conditions.
- AFM Imaging and Abrasion:
- Image a 5x5 μm area of the biofilm at a low applied load (~0 nN) to obtain a baseline topography.
- Zoom into a 2.5x2.5 μm sub-region and perform four consecutive raster scans at a high load (40 nN) to abrade the biofilm.
- Return to a low load and image the same 5x5 μm area again to capture the abrasion scar.
- Data Analysis:
- Subtract the post-abrasion image from the pre-abrasion image to calculate the volume of biofilm displaced.
- Use the friction signal recorded during abrasive scanning to calculate the frictional energy dissipated.
- Calculate the cohesive energy as the frictional energy divided by the volume of displaced biofilm.
The Scientist's Toolkit
Table 2: Essential Research Reagents and Materials
| Item | Function / Application | Example Usage |
|---|---|---|
| PFOTS-treated Glass | Creates a hydrophobic surface for studying initial bacterial attachment and biofilm assembly [3]. | Used as a substrate for Pantoea sp. YR343 attachment in large-area AFM studies [3]. |
| COOH-functionalized Polystyrene Beads | Serve as carriers for biofilm growth; suitable for bacterial adhesion and compatible with FluidFM [43]. | Beads are colonized by biofilms and then aspirated onto a FluidFM cantilever for adhesion force measurements [43]. |
| Recombinant Human SIRPα1 | A prototypical ligand used to functionalize AFM tips for specific receptor-binding studies [45]. | AFM tips coated with SIRPα are used to probe its native receptor, CD47, on red blood cell membranes [45]. |
| Vanillin-coated Membranes | Act as an anti-biofouling surface; vanillin is a quorum-sensing inhibitor that reduces EPS production [43]. | Used as a test surface in FluidFM experiments to quantify the reduction in biofilm adhesion forces [43]. |
| Silanized AFM Tips | Provides a chemically active surface for stable immobilization of proteins or other biomolecules [45]. | Tips are silanized with Allyltrichlorosilane before functionalization with SIRPα for specific adhesion experiments [45]. |
Experimental Workflow and Data Analysis
The diagram below outlines a logical workflow for addressing AFM artifacts in biofilm research, integrating the strategies discussed.
Frequently Asked Questions
1. What are the most common data processing artifacts in AFM of biofilms, and how can I identify them? The most common artifacts arise from tip convolution, improper flattening, and excessive filtering. Tip convolution causes fine features like flagella to appear wider and makes narrow gaps between cells seem smaller or shallower [3] [46]. Over-flattening can create a falsely smooth surface, removing critical height information and masking the true roughness and heterogeneity of the biofilm [46]. Excessive filtering smears out real topographical features, and hysteresis of piezoelectric scanner elements can cause distortions in the X-Y plane unless corrected with modern closed-loop systems or software [46].
2. My AFM images of bacterial cells lack the expected detail. Is this a data processing or hardware issue? It can be both. The physical AFM tip is a primary limitation; a tip with a low aspect ratio or a blunt tip will physically fail to resolve narrow gaps or fine structures like flagella, a problem known as tip convolution [46]. No amount of data processing can recover information that was never physically recorded. During processing, applying a filter that is too strong can erase these same fine details. Always inspect raw data before processing and ensure your AFM tip is sharp and suitable for your sample's topography.
3. How does sample heterogeneity in biofilms complicate data flattening? Standard flattening algorithms assume a relatively uniform background. Biofilms are intrinsically heterogeneous, with a mix of cells, extracellular polymeric substances (EPS), and voids [3]. If you select a flattening line or plane that crosses a very high feature (like a cell cluster) and a very low area (a void), the algorithm will incorrectly "tilt" the entire image, distorting the accurate height of all other features. The resulting image may look flat, but the quantitative height data will be unreliable.
4. What are the best practices for filtering AFM data on heterogeneous biofilm samples? The best practice is a minimal and informed approach. Always keep a copy of the raw, unprocessed data. Apply filters sparingly and only after a correct flattening procedure. Use the mildest possible filter strength that reduces high-frequency noise without blurring legitimate biological structures. Compare the filtered image side-by-side with the raw data to ensure critical features are preserved. Document all processing steps and parameters used for future reproducibility.
5. Can AI and machine learning help avoid these pitfalls? Yes, AI and machine learning are transforming this field. Machine learning models can now automate the distinction between true surface features and common artifacts, leading to more objective analysis [3] [11]. For highly heterogeneous samples, AI-driven automated scanning can optimize data acquisition across millimeter-scale areas, ensuring representative sampling [3]. Furthermore, new software like AFMfit uses flexible fitting procedures to help interpret the conformational states of molecules in AFM images, reducing model-based misinterpretation [47].
Troubleshooting Guides
Problem 1: Loss of Surface Roughness and Fine Features After Flattening
- Symptoms: The biofilm surface appears artificially smooth; fine structures like flagella, pili, or EPS fibers are faint or missing; quantitative roughness measurements (e.g., Rq, Ra) are lower than expected.
- Root Cause: The flattening algorithm was applied too aggressively or an inappropriate reference plane was selected. On heterogeneous samples, using a single global flattening function can "flatten out" real topographic variation.
- Solution:
- Re-process with care: Use a line-by-line flattening (0th or 1st order) instead of a higher-order global plane fit.
- Leverage AI tools: Implement machine learning-based segmentation to identify and mask large cell clusters before flattening, allowing the algorithm to fit only to the background surface [11].
- Validate quantitatively: Compare the roughness parameters of the raw data (after minimal, correct flattening) with the over-processed data to quantify the information loss.
Problem 2: Artificial "Softening" or "Smearing" of Cell Edges
- Symptoms: Bacterial cells look rounded and lack defined edges; the image appears out-of-focus or blurry.
- Root Cause: This is a classic sign of tip convolution or excessive low-pass filtering [46]. The physical size and shape of the AFM tip are too large to probe the steep sides of the cells accurately, and subsequent filtering exacerbates the problem.
- Solution:
- Tip Selection: Use AFM tips with a high aspect ratio and sharp apex for imaging complex biofilm architectures [46].
- Filter Judiciously: Avoid strong low-pass filters. If necessary, use a mild filter and compare with the raw data to ensure edge sharpness is maintained.
- Deconvolution: Apply tip-deconvolution algorithms that use a model of the tip's shape to reconstruct a more accurate image of the surface, though this requires knowing your tip's geometry [47].
Problem 3: Inconsistent Height Measurements Across an Image
- Symptoms: The measured height of identical bacterial cells varies significantly depending on their location in the scan area.
- Root Cause: This is often due to scanner hysteresis or non-linearity in the piezoelectric elements, or an improper flattening that used an uneven baseline [46].
- Solution:
- Scanner Maintenance: Use a modern AFM with a closed-loop scanner, which actively corrects for hysteresis and cross-talk, virtually eliminating this problem [46].
- Calibration: Regularly calibrate the scanner using a reference grating with known dimensions.
- Flattening Check: Ensure flattening is performed on a stable, featureless region of the substrate, not on the biofilm itself.
Data Presentation
Table 1: Common AFM Data Processing Artifacts and Their Impact on Biofilm Interpretation
| Artifact Type | Cause | Effect on Image | Impact on Biofilm Research |
|---|---|---|---|
| Tip Convolution [46] | Physical size/shape of AFM tip limits resolution. | Fine structures (flagella) appear wider; narrow gaps are obscured. | Mischaracterization of cell appendages and cell-cell interaction space. |
| Over-Flattening [46] | Overly aggressive application of flattening algorithms. | Artificial smoothing; loss of surface roughness and height data. | False quantification of biofilm architecture and heterogeneity. |
| Excessive Filtering [46] | Application of strong noise-reduction filters. | Loss of high-resolution detail; "smearing" of edges. | Inaccurate measurement of cell dimensions and EPS matrix structure. |
| Scanner Hysteresis [46] | Non-linear movement of piezoelectric scanner. | Distortions in X-Y plane; inconsistent feature sizes/locations. | Incorrect spatial mapping of cell clusters and community organization. |
Table 2: Quantitative Guide to Filter Selection for Biofilm Features
| Biofilm Feature | Typical Size Scale | Recommended Filter Type | Caution |
|---|---|---|---|
| Flagella / Pili [3] | 20-50 nm diameter | Very mild low-pass or plane fit only | Highly susceptible to blurring; avoid filtering if possible. |
| Individual Cells [3] | 1-2 µm (diameter) | Mild low-pass or flattening with 1st order | Preserve sharp edges for accurate dimensioning. |
| EPS Matrix | >50 nm (variable) | Mild low-pass | Goal is to reduce graininess without losing fibrous texture. |
| Cell Clusters [3] | 5-100 µm | Plane fit or flattening | Ensure flattening does not use cluster as reference. |
Experimental Protocols
Protocol 1: Minimizing Artifacts During AFM Imaging of Biofilms (Adapted from [3])
- Sample Preparation: Grow Pantoea sp. YR343 biofilm on PFOTS-treated glass coverslips. Gently rinse with buffer to remove non-adherent cells. For imaging in air, allow to dry.
- Tip Selection: Choose a sharp, high-aspect-ratio silicon tip to minimize convolution artifacts [46].
- Initial Scan:
- Use a large scan size (e.g., 50x50 µm) to identify areas of interest and assess sample heterogeneity.
- Set a slow scan rate (e.g., 0.5-1 Hz) to improve signal-to-noise ratio.
- High-Resolution Scan:
- Zoom into a representative region (e.g., 10x10 µm).
- Increase the resolution (512x512 or 1024x1024 pixels).
- Ensure the setpoint and gains are optimized to maintain tip contact without damaging the sample.
- Data Saving: Save a copy of the raw, unprocessed data file before any post-processing.
Protocol 2: A Robust Data Processing Workflow for Heterogeneous Biofilms
- Raw Data Inspection: Always begin by visually inspecting the raw data for major scanner artifacts or large-scale tilts.
- Minimal Flattening:
- Apply a 0th or 1st order flattening function.
- Select a reference line that lies on the flat substrate, avoiding cells and large aggregates. For large-area scans, use a multi-point plane fit if available.
- Noise Reduction:
- Apply a light low-pass filter or a 2D FFT filter to remove high-frequency noise.
- The filter strength should be just enough to reduce pixel noise without blurring cell edges or fine filaments.
- Analysis:
The Scientist's Toolkit
Table 3: Research Reagent Solutions for AFM of Biofilms
| Item | Function in Experiment | Example & Notes |
|---|---|---|
| PFOTS-treated Glass [3] | Creates a hydrophobic surface to promote controlled bacterial adhesion for consistent AFM sample preparation. | Used in Pantoea sp. YR343 studies to observe early-stage biofilm formation [3]. |
| High-Aspect-Ratio AFM Tips [46] | Sharp, long tips are essential for accurately probing the deep crevices and steep sides of features in a mature, heterogeneous biofilm. | Critical for minimizing tip-convolution artifacts and obtaining true topographic data. |
| AI-Powered Image Analysis Software [3] [11] [47] | Automates stitching of large-area scans, segments individual cells from EPS, and classifies features, reducing subjective bias in analysis. | Tools like large-area automated AFM with ML stitching and AFMfit for conformational analysis are becoming standard [3] [47]. |
| Closed-Loop AFM Scanner [46] | Actively corrects for piezoelectric hysteresis and drift, providing real-time, accurate positioning and eliminating spatial distortion artifacts. | Essential for achieving precise, quantifiable measurements across large scan sizes. |
Workflow Visualization
Correct AFM Data Processing Workflow
Artifact Generation from Tip & Filtering
Validating AFM Data: Correlative Microscopy and Comparative Analysis for Biofilm Studies
Biofilms are complex, heterogeneous microbial communities where structure and function are deeply intertwined. A single imaging technique often fails to capture the complete picture, as each method has inherent limitations and strengths. Atomic Force Microscopy (AFM) provides exceptional nanoscale topographical and mechanical data but lacks chemical specificity and can be limited in scan area. Correlative microscopy addresses these limitations by integrating AFM with complementary techniques like Confocal Laser Scanning Microscopy (CLSM), Scanning Electron Microscopy (SEM), and Raman Spectroscopy. This synergy allows researchers to cross-validate findings, providing a more comprehensive and accurate understanding of biofilm architecture, composition, and function, which is crucial for developing effective control strategies.
The following sections provide a technical support framework, offering troubleshooting guides and detailed protocols to successfully implement this powerful correlative approach in your biofilm research.
Troubleshooting Guide: Resolving Common Correlative Workflow Challenges
FAQ: What are the most frequent pitfalls when correlating AFM with other imaging techniques, and how can they be avoided?
Integrating different microscopy platforms often presents challenges related to sample compatibility, data alignment, and artifact interpretation. The table below summarizes common issues and their solutions.
Table 1: Troubleshooting Common Issues in Correlative Microscopy for Biofilms
| Problem | Potential Cause | Solution | Preventive Measure |
|---|---|---|---|
| Poor spatial correlation between AFM and optical/CLSM images. | Lack of distinct, permanent fiduciary markers on the substrate. | Use substrates with pre-fabricated coordinate grids or deposit microbeads as landmarks. | Plan the correlative workflow before sample preparation; select appropriate marked substrates. |
| AFM tip contamination or sample damage during scanning. | The biofilm's extracellular polymeric substance (EPS) is sticky and soft. | Operate AFM in tapping mode in liquid to minimize shear forces [48]. | Engage the AFM tip gently and use sharper, high-frequency probes for softer samples. |
| Inconsistent findings between AFM topography and Raman chemical maps. | The Raman signal is averaged over a larger volume than AFM tip contact. | Ensure the AFM tip radius is comparable to the Raman laser spot size, or use tip-enhanced Raman spectroscopy (TERS). | Acknowledge the scale difference; use multivariate analysis (e.g., PCA) on Raman data for better feature isolation [49]. |
| Sample deformation between SEM and AFM analysis. | SEM sample preparation (dehydration, metal coating) alters native biofilm structure. | Perform AFM and Raman first to analyze the hydrated, native state. If possible, use environmental SEM (ESEM) to minimize dehydration. | Establish a workflow order that prioritizes non-destructive techniques (CLSM, Raman, AFM) before destructive ones (SEM). |
| Low signal-to-noise ratio in Raman spectra from biofilms. | High fluorescence background from EPS or culture media, weak Raman scattering. | Implement rigorous background correction and spectral calibration [50]. Use surface-enhanced Raman scattering (SERS) if higher sensitivity is needed. | Employ a laser wavelength in the near-infrared (e.g., 785 nm) to reduce fluorescence and ensure proper intensity calibration [50]. |
Detailed Experimental Protocols for Cross-Validation
Protocol: Correlating AFM with CLSM for Structural and Viability Analysis
This protocol is designed to link the nanoscale surface properties measured by AFM with the 3D internal structure and cell viability information from CLSM.
Key Research Reagent Solutions:
- Fluorescent Stains: SYTO 9 (for live-cell nucleic acids), Propidium Iodide (for dead-cell nucleic acids), Concanavalin A conjugated with a fluorescent dye (for labelling EPS polysaccharides).
- Substrate: Glass-bottom Petri dishes or WillCo-dishes, ideal for high-resolution optical microscopy and AFM.
- Buffer: A suitable physiological buffer (e.g., PBS) to maintain biofilm viability during imaging.
Methodology:
- Sample Preparation: Grow a biofilm on a glass-bottom dish. Gently rinse with buffer to remove non-adherent cells.
- Staining: Incubate the biofilm with the chosen fluorescent dyes (e.g., LIVE/DEAD BacLight bacterial viability kit) according to the manufacturer's protocol. Avoid excessive light exposure.
- CLSM Imaging: First, acquire Z-stacks of the fluorescently labelled biofilm using CLSM. Identify and document regions of interest (ROIs) based on structural features or viability patterns. Capture high-resolution optical images of these ROIs, noting the coordinates.
- Transfer to AFM: Carefully transfer the dish to the AFM stage, ensuring the liquid environment is maintained.
- AFM Imaging: Use the optical microscope integrated with the AFM to relocate the pre-defined ROIs. Engage the AFM tip and perform scanning in tapping mode in liquid. Acquire high-resolution topographical images and, if applicable, nanomechanical property maps (e.g., Young's modulus) of the same ROIs.
- Data Correlation: Overlay the AFM topography map with the CLSM fluorescence image using the fiduciary markers and distinct cellular features for precise alignment. This allows direct comparison of, for instance, the height of a microcolony (AFM) with the distribution of live/dead cells within it (CLSM).
Protocol: Integrating AFM with Raman Spectroscopy for Chemo-Mechanical Mapping
This protocol leverages the strength of AFM to map physical properties and Raman to provide a molecular "fingerprint," enabling the correlation of structure with biochemistry.
Methodology:
- Sample Preparation: Grow a biofilm on an optimized surface (e.g., silicon wafer or PFOTS-treated glass [3]). Gently rinse and air-dry if required for the specific Raman measurement, though hydrated measurement is preferable.
- AFM Analysis: Perform an initial large-area scan (if available, using an automated large-area AFM [3]) to identify areas with heterogeneous features.
- Raman Analysis: Transfer the sample to the Raman microscope. Relocate the ROIs analyzed by AFM. Acquire Raman spectra from a grid of points across the ROI to create a hyperspectral image.
- Data Analysis Pipeline for Raman:
- Preprocessing: Correct for cosmic rays.
- Calibration: Perform wavenumber and intensity calibration. Critical Note: Avoid the mistake of performing spectral normalization before background correction, as this can bias the results [50].
- Background Correction: Subtract the fluorescent baseline from the raw spectra.
- Multivariate Analysis: Use Principal Component Analysis (PCA) or Cluster Analysis (CA) to differentiate spectral types, which can be mapped back to the spatial coordinates to identify chemical domains [49].
- Data Correlation: Correlate the AFM-derived topography and stiffness maps with the Raman chemical maps. For example, stiff, dense clusters of cells observed in AFM can be validated against Raman maps showing high nucleic acid and protein signals, while softer, amorphous areas might correlate with EPS-specific Raman bands.
Table 2: Key Reagents and Their Functions in Correlative Biofilm Studies
| Reagent/Material | Function in Experiment |
|---|---|
| PFOTS-treated glass coverslips | Creates a hydrophobic surface to study specific bacterial adhesion and biofilm assembly patterns [3]. |
| SYTO 9 / Propidium Iodide | Fluorescent viability stains for CLSM to distinguish between live and dead bacterial populations within the biofilm. |
| Silicon or Silicon Nitride AFM Probes | Sharp tips (nominal radius <10 nm) for high-resolution topographical and mechanical mapping of delicate biofilm surfaces. |
| 4-Acetamidophenol | A wavenumber standard used for the critical calibration of the Raman spectrometer, ensuring spectral accuracy and reproducibility [50]. |
| Brain Heart Infusion (BHI) Medium | A rich growth medium used for cultivating model biofilm organisms like Streptococcus oralis and Actinomyces denticolens [49]. |
Workflow Visualization: The Integrated Correlative Pathway
The following diagram illustrates the optimal workflow for a fully integrated correlative study, from sample preparation to data synthesis, highlighting the sequence of techniques to maximize information recovery.
Advanced Applications: Leveraging Machine Learning and Large-Area AFM
The volume and complexity of data generated by correlative microscopy necessitate advanced analysis tools. Machine Learning (ML) and Artificial Intelligence (AI) are now transforming this field.
- Automated Image Analysis: ML algorithms can automate the segmentation of individual cells and EPS from AFM topographical images or CLSM stacks, enabling high-throughput quantification of parameters like cell count, confluency, and shape orientation [3] [11].
- Enhanced Raman Data Interpretation: AI-based data analysis pipelines are crucial for correcting, standardizing, and interpreting Raman spectra. Deep learning models can translate spectral "fingerprints" into high-level information about biofilm composition and function [11] [50].
- Large-Area AFM: Traditional AFM's limited scan area is a major constraint. New automated large-area AFM approaches, aided by ML for image stitching, can now capture high-resolution images over millimeter-scale areas. This directly bridges the gap between the macro-scale organization visible to CLSM and the nanoscale features resolved by standard AFM, making correlation far more representative and powerful [3].
The integration of AFM with SEM, CLSM, and Raman spectroscopy represents a paradigm shift in biofilm research. This correlative approach moves beyond the limitations of any single technique, enabling robust cross-validation and a multidimensional understanding of biofilm systems. By adhering to detailed protocols for sample preparation, workflow sequencing, and data analysis—and by leveraging new capabilities in machine learning—researchers can minimize artifacts and unlock new insights. As these technologies continue to converge and advance, they promise to redefine the landscape of biofilm research and accelerate the development of novel anti-biofilm strategies.
Atomic Force Microscopy (AFM) provides powerful topographical data for biofilm research, enabling nanoscale resolution of cellular morphology and extracellular polymeric substances. However, the inherent heterogeneity of biofilms and the prevalence of AFM imaging artifacts necessitate rigorous benchmarking against established biological methods. This technical support center provides troubleshooting guidance and protocols for researchers validating AFM data against conventional staining and colony forming unit (CFU) approaches, ensuring accurate interpretation of structural and mechanical properties in complex biofilm samples.
Troubleshooting Guide: Resolving Common AFM Benchmarking Challenges
FAQ 1: How do I resolve discrepancies between AFM cell counts and CFU measurements?
Issue: Significant differences observed between bacterial densities quantified via AFM topography and traditional CFU plating.
Solution:
- Investigate cellular viability state: CFU assays only detect culturable cells, while AFM counts all surface-attached cells, including dormant or viable-but-non-culturable (VBNC) states. Supplement with viability staining (e.g., LIVE/DEAD) to resolve [51].
- Verify biofilm dispersal method: Inadequate dissociation of biofilm aggregates during CFU preparation underestimates counts. Optimize homogenization (e.g., vortexing with glass beads, enzymatic matrix degradation) [52].
- Account for AFM sampling limitations: Small AFM scan areas may not represent heterogeneous biofilm architecture. Implement large-area AFM mapping with automated cell detection to improve statistical significance [3].
Prevention Protocol:
- Correlate AFM data with peptide nucleic acid fluorescence in situ hybridization (PNA-FISH) for accurate quantification of total cells regardless of viability [51].
- Standardize biofilm growth conditions and sampling locations across all analysis methods.
FAQ 2: What causes poor correlation between EPS visualization in AFM and fluorescent staining?
Issue: Extracellular polymeric substance (EPS) distribution patterns differ between AFM topographical maps and fluorescent conjugate stains (e.g., WGA, concanavalin A).
Solution:
- Identify staining specificity limitations: Fluorescent probes may bind non-specifically to host tissue components or fail to penetrate dense biofilm matrix, yielding false positives/negatives [51].
- Optimize AFM operational mode: Use quantitative nanomechanical mapping modes to distinguish EPS from cellular components based on mechanical properties (adhesion, deformation) rather than topography alone.
- Control sample dehydration effects: Air-drying for AFM can collapse hydrogel-like EPS, altering native architecture. Compare with environmental AFM under physiological buffers [53].
Prevention Protocol:
- Validate staining protocols with known EPS component controls.
- Fix samples with 2.5% glutaraldehyde for optimal preservation of surface ultrastructures before either AFM or staining procedures [53].
FAQ 3: How do I minimize artifacts when imaging heterogeneous biofilm topography?
Issue: AFM images show features that may represent imaging artifacts rather than true biofilm structures, complicating data interpretation.
Solution:
- Characterize tip geometry: Regularly image reference nanostructures (e.g., TED Pella grid) to monitor tip sharpness and identify tip convolution effects [54].
- Optimize scanning parameters: For soft biofilm samples, reduce tapping force, increase oscillation amplitude, and lower scan rates to minimize sample deformation and trailing artifacts [55] [54].
- Implement drift compensation: Use closed-loop scanners and thermal equilibrium protocols to minimize spatial distortions during long-duration scans of large biofilm areas [3] [54].
Prevention Protocol:
- Employ machine learning-enhanced AFM systems that automatically optimize scanning parameters and identify common artifact patterns [3] [31].
- Always image multiple locations and compare with secondary imaging techniques (e.g., SEM) to confirm key structural findings.
Method Comparison Framework: Quantitative Analysis of Complementary Techniques
Table 1: Comparison of Biofilm Characterization Techniques: Capabilities and Limitations
| Method | Resolution | Information Obtained | Key Limitations | Best Applications |
|---|---|---|---|---|
| AFM Topography | 0.5-1 nm vertical, 10-30 nm lateral [3] | 3D surface morphology, nanomechanical properties, surface roughness | Limited field of view, potential sample deformation, tip artifacts | Single-cell morphology, EPS nanoscale organization, surface adhesion forces |
| CFU Counting | N/A | Viable, culturable cell counts | Misses VBNC states, requires biofilm disruption, 24-48 hour delay | Antimicrobial efficacy testing, quantifying viable bacterial load |
| Fluorescent Staining | ~200-300 nm (diffraction-limited) | Chemical composition, viability, specific molecule distribution | Dye penetration issues, photobleaching, non-specific binding | Spatial organization of living/dead cells, specific EPS component identification |
| SEM Imaging | 1-10 nm [51] | High-resolution surface architecture, biofilm-host tissue interface | Requires dehydration/coating, no live imaging, no mechanical data | Visualizing biofilm aggregation in complex wound tissue [51] |
Table 2: Troubleshooting AFM Artifacts in Biofilm Research
| Artifact Type | Common Causes | Impact on Data | Corrective Actions |
|---|---|---|---|
| Tip Convolution | Blunt/damaged probes, inappropriate tip geometry | Overestimation of feature widths, loss of fine details | Use sharper tips (radius <10 nm), regular tip characterization, deconvolution algorithms [53] |
| Thermal Drift | Temperature fluctuations during scanning | Image distortion, spatial inaccuracies | Allow scanner thermal equilibrium, use drift compensation, shorten scan times |
| Feedback Overshoot | Improper PID settings on rough surfaces | "Shadow" effects, ringing near edges | Reduce scan speed, optimize gains, use adaptive feedback systems [56] |
| Sample Deformation | Excessive imaging force on soft biofilms | Compressed features, altered morphology | Use softer cantilevers (0.1-2 N/m), reduce engagement force, employ tapping mode [55] |
Experimental Protocols for Method Validation
Protocol 1: Correlative AFM and Fluorescence Imaging for EPS Validation
Sample Preparation:
- Grow biofilms on sterile glass coverslips suitable for both microscopy techniques.
- Fix with 2.5% glutaraldehyde in PBS for 2 hours at room temperature for structural preservation [53].
- Rinse gently with PBS and divide samples for parallel processing.
AFM Imaging:
- Use tapping mode with soft cantilevers (spring constant: 0.1-2 N/m) in liquid if possible.
- Capture large-area scans (≥50×50 μm²) followed by high-resolution zooms (5×5 μm²) on regions of interest.
- Acquire both height and phase channels to distinguish material properties.
Fluorescence Staining:
- Apply appropriate fluorescent conjugates (e.g., WGA for polysaccharides, SYPRO Ruby for proteins).
- Image identical regions using confocal microscopy with minimal laser power to prevent photobleaching.
- Use image registration software to align AFM and fluorescence datasets.
Data Correlation:
- Overlay AFM topography with fluorescence channels to verify spatial correspondence.
- Quantify colocalization using Pearson's correlation coefficient for objective comparison.
Protocol 2: Integrated AFM-CFU Analysis for Antimicrobial Efficacy Testing
Biofilm Treatment:
- Grow standardized biofilms (e.g., 24-48 hours) and apply antimicrobial treatment.
- Prepare triplicate samples for each analysis method from the same biofilm culture.
AFM Quantification:
- Use large-area AFM with automated image stitching to scan multiple regions [3].
- Apply machine learning-based segmentation to identify and count individual cells across mm² areas.
- Calculate surface coverage (%) and average cell height as structural metrics.
CFU Enumeration:
- Harvest biofilm cells from identical treatment conditions using established scraping/sonication protocols.
- Perform serial dilution and plating on appropriate agar media.
- Incubate and count colonies after 24-48 hours.
Data Normalization:
- Express both datasets as percentage reduction compared to untreated controls.
- Account for the difference between total cells (AFM) and viable cells (CFU) in data interpretation.
Experimental Workflow and Logical Relationships
Research Reagent Solutions: Essential Materials for AFM Biofilm Studies
Table 3: Key Research Reagents and Materials for AFM Biofilm Studies
| Reagent/Material | Function | Application Notes | References |
|---|---|---|---|
| Glutaraldehyde (2.5%) | Additive fixation preserving surface ultrastructures | Superior to alcohol fixatives for flagella, pili, and EPS visualization | [53] |
| Silicon Cantilevers | AFM probes for topography imaging | Use soft levers (0.1-2 N/m) for biofilms; Tap300 for high resolution | [53] |
| PFOTS-treated Glass | Hydrophobic substrate for controlled biofilm growth | Promotes specific cellular orientation patterns in Pantoea sp. | [3] |
| WGA Conjugates | Fluorescent staining of EPS polysaccharides | Limited specificity in complex host tissue; requires validation | [51] |
| PNA-FISH Probes | Molecular identification of biofilm bacteria | Gold standard validation for AFM cell counts, detects VBNC cells | [51] |
| HMDS | Sample dehydration for SEM correlation | Prevents structural collapse during drying process | [51] |
| Calibration Gratings | AFM scanner calibration and tip characterization | Essential for quantifying and minimizing measurement artifacts | [54] |
Technical FAQs: Bridging AFM and Bulk Rheology
FAQ 1: What are the primary causes of discrepancy between nanomechanical AFM maps and bulk rheology data for biofilms?
Discrepancies often arise from fundamental differences in what each technique measures. AFM probes local, nanoscale properties of a biofilm's heterogeneous structure, such as individual cells and the surrounding extracellular polymeric substance (EPS). In contrast, bulk rheology provides the average macroscopic response of the entire biofilm sample. Key causes for disagreement include:
- Spatial Heterogeneity: AFM might sample a stiff bacterial cell in one measurement and a soft EPS region in another, while rheology averages these extremes. The inherent spatial heterogeneity of biofilms is a significant factor that can obscure direct correlation [3].
- Probe-Sample Interactions: AFM measurements can be influenced by adhesion forces between the tip and the sample, which are not a factor in bulk rheology [57] [58].
- Strain and Stress Fields: The strain field under an AFM tip is highly localized and nonlinear, whereas bulk rheology typically applies a uniform, well-defined stress or strain [58].
- Hydration State: AFM can be performed under physiological liquid conditions, but some protocols may require partial drying, which dramatically alters mechanical properties. Bulk rheology is typically performed in a fully hydrated state [29].
FAQ 2: Which AFM modes are most suitable for collecting data that can be correlated with bulk rheology?
For meaningful correlation with bulk rheology, which often measures viscoelastic properties, AFM modes that can quantify both elastic and viscous components are preferred.
- Force Volume Mode: This mode involves acquiring a force-distance curve on each pixel of the image. It allows for the calculation of elasticity (Young's modulus) and adhesion forces. The hysteresis observed between the approach and retraction curves can provide insights into energy dissipation, a key viscoelastic parameter [58].
- Nano-DMA (Dynamic Mechanical Analysis) Modes: These are the most direct analogues to bulk rheology. The AFM tip is held in contact with the sample and an oscillatory signal is applied. By analyzing the amplitude and phase lag of the tip's response, one can map storage modulus (elasticity) and loss modulus (viscosity) [58].
- Force Modulation Mode: This mode is used to measure the elasticity of the sample by applying a high-frequency oscillation to the tip or sample while in contact and monitoring the cantilever's response [59].
Table 1: Comparison of AFM Modes for Nanomechanical Mapping
| AFM Mode | Measured Parameters | Advantages | Considerations for Correlation with Bulk Rheology |
|---|---|---|---|
| Force Volume | Young's Modulus, Adhesion, Deformation | Direct force measurement; can detect viscoelasticity via hysteresis | Slow data acquisition; point-by-point analysis required |
| Nano-DMA | Storage & Loss Moduli, Tan Delta | Directly measures viscoelasticity; similar principles to bulk DMA | Requires precise calibration; can be sensitive to drift |
| Force Modulation | Relative Stiffness | Good for qualitative stiffness mapping | Less quantitative for absolute modulus values compared to force volume |
| PeakForce QI | Young's Modulus, Adhesion, Dissipation | High-speed force mapping; reduces sample damage | Proprietary implementation (Bruker) |
FAQ 3: How can we troubleshoot common artifacts in AFM nanomechanical maps of soft, heterogeneous biofilms?
AFM imaging of soft biofilms is prone to artifacts that can compromise data validity and correlation efforts.
Artifact: "Stripe" or "Scan Line" Patterns
- Cause: Poor feedback loop optimization, especially when scanning from a stiff substrate to a very soft biofilm.
- Solution: Optimize feedback gains (proportional and integral) to ensure the tip can track the surface accurately without oscillating. Reduce the scan rate [25].
Artifact: Unrealistically High Modulus Values at Cell Margins
- Cause: This is often a "tip-convolution" effect. The finite size and shape of the tip cannot properly resolve the steep edges of cells, leading to an overestimation of stiffness.
- Solution: Use sharper, high-resolution tips. Apply deconvolution algorithms during data processing if available [25] [60].
Artifact: Apparent Changes in Topography and Mechanics from Repeated Scanning
- Cause: The AFM tip is physically displacing or damaging the soft biofilm material.
- Solution: Reduce the applied force (setpoint). Use a softer cantilever (lower spring constant) appropriate for soft samples. Validate results by scanning the same area multiple times to check for consistency [29].
Artifact: Inconsistent Measurements on the Same Sample
- Cause: The biofilm's inherent heterogeneity means different scan areas have different compositions.
- Solution: Implement large-area AFM scanning by automatically stitching multiple high-resolution images together. This provides a more representative dataset for comparison with bulk techniques [3].
Experimental Protocol: Cohesive Energy Measurement for Biofilm Stability
This protocol, adapted from a foundational study, details how to use AFM to measure biofilm cohesive energy, a critical parameter for understanding stability and detachment that can be related to bulk strength [29].
Goal: To measure the cohesive energy (nJ/μm³) of a moist biofilm in situ via AFM-based abrasion.
Key Research Reagent Solutions:
- Biofilm Support: Microporous polyolefin flat sheet membrane (e.g., 3M Corporation) [29].
- Immobilization Substrate: Freshly cleaved mica disks for high-resolution imaging of bacterial cells [60].
- Cantilever Selection: V-shaped silicon nitride cantilevers with oxide-sharpened pyramidal tips (e.g., Bruker NP-S). A spring constant of ~0.58 N/m is suitable [29].
- Hydration Control: A saturated NaCl solution in a closed chamber to maintain a constant humidity of ~90% during imaging, preserving the biofilm's native state [29].
Step-by-Step Methodology:
- Biofilm Growth and Preparation: Grow a 1-day-old biofilm on a membrane in a reactor. Cut a ~1 cm² sample and equilibrate it in a 90% relative humidity chamber for one hour to maintain consistent water content.
- Baseline Topographic Imaging: Mount the hydrated biofilm sample in the AFM. On a 5x5 μm area of interest, collect a non-perturbative topographic image using a minimal applied force (∼0 nN). Record this "before" image.
- Controlled Abrasion Scanning: Zoom into a 2.5x2.5 μm sub-region within the previously scanned area. Set a high applied load (e.g., 40 nN) and perform repeated raster scans (e.g., 4 scans) to abrade the biofilm surface.
- Post-Abrasion Topographic Imaging: Return to the low applied load and capture another non-perturbative 5x5 μm image of the abraded region. This is the "after" image.
- Data Analysis:
- Volume Loss Calculation: Subtract the "after" topographic image from the "before" image using AFM software. Calculate the total volume (μm³) of biofilm displaced by the abrasion.
- Frictional Energy Calculation: During the abrasive scanning, the lateral deflection signal (friction force) is recorded. Integrate this friction force over the total scan distance to calculate the total frictional energy dissipated (nJ).
- Cohesive Energy Calculation: Divide the total frictional energy by the volume of displaced biofilm. The result is the cohesive energy density in nJ/μm³.
Experimental Workflow for AFM Cohesive Energy Measurement
Data Correlation Framework: From Nanoscale to Macroscale
Successfully correlating AFM and bulk rheology data requires a systematic approach to account for the different scales and physical principles involved.
Table 2: Framework for Correlating AFM and Bulk Rheology Data
| Aspect | AFM Nanomechanics | Bulk Rheology | Correlation Strategy |
|---|---|---|---|
| Spatial Scale | Nanometers to Micrometers (local) | Millimeters (global average) | Perform large-area AFM mapping to compute spatial averages and distributions of properties [3]. |
| Probed Volume | Femtoliters to Picoliters | Microliters to Milliliters | Use AFM to intentionally map multiple structural components (cells, EPS) and create a weighted average model. |
| Measured Properties | Young's Modulus, Adhesion, Cohesive Energy | Storage/Loss Modulus, Yield Stress, Complex Viscosity | Focus on trends rather than absolute values. For example, correlate how AFM adhesion and cohesive energy scale with bulk yield stress [29]. |
| Sample Preparation | Can be done in liquid or controlled humidity; may require surface immobilization. | Typically fully hydrated in a sealed geometry. | Ensure hydration states are as similar as possible. AFM in liquid is preferred for biological relevance [59]. |
| Data Output | Maps of properties (images), force curves. | Averaged numbers, flow curves, frequency sweeps. | Extract statistical parameters (mean, median, standard deviation) from AFM maps for direct comparison with rheological averages. |
AFM and Rheology Data Correlation Logic
Assessing Reproducibility and Statistical Significance in Heterogeneous Biofilm Samples
Troubleshooting Guides
Guide 1: Addressing Atomic Force Microscopy (AFM) Artifacts in Biofilm Imaging
Problem: AFM images of heterogeneous biofilms show inconsistencies or features suspected to be artifacts, not true biological structures.
- Potential Cause 1: Tip-Sample Interaction Artifacts. The AFM tip physically deforms or damages the soft, heterogeneous biofilm surface during scanning.
- Solution: Switch from contact mode to tapping (intermittent contact) mode. This reduces lateral forces and minimizes damage to soft samples, leading to higher image accuracy [61]. Ensure the cantilever's oscillation amplitude and setpoint are optimized for the biofilm's stiffness.
- Potential Cause 2: Poor Tip Quality. A contaminated, worn, or broken AFM tip produces repeating, exaggerated, or distorted features.
- Solution: Regularly inspect tips using a reference sample with known morphology. Clean the tip according to manufacturer protocols and replace it if damage is suspected. The system's high magnification and sensitivity should be used to identify and rectify tip-related artifacts [61].
- Potential Cause 3: Improper Sample Preparation. Cells are detached during scanning, or chemical fixation alters the biofilm's native mechanical properties.
- Solution: For live cell imaging, carefully immobilize the biofilm on a suitable substrate under physiological conditions (e.g., in nutrient medium with controlled temperature). Avoid chemical fixation where possible, as it can make cells stiffer and introduce artifacts. If cells detach, find an intact section or prepare a new sample [61].
- Potential Cause 4: Scanner Calibration Errors. Inaccurate movement of the piezoelectric scanner leads to distorted spatial measurements.
- Solution: Regularly calibrate the AFM scanner using a calibration grating with features of known dimensions. This ensures accurate measurements of biofilm topographic features like height and volume [61].
Problem: Low reproducibility in nanomechanical measurements (e.g., stiffness, adhesion) across different biofilm samples.
- Potential Cause: Lack of Standardized Criteria. Inconsistent measurement parameters (e.g., indentation force, speed, location) and analysis models make cross-study comparisons difficult.
- Solution: Establish and rigorously document a standard operating procedure (SOP) for your lab. This should include:
- Cantilever Calibration: Precisely determine the spring constant of each cantilever before use.
- Measurement Location: Define a systematic grid or pattern for indentation points to account for heterogeneity.
- Analysis Model: Consistently use the same mechanical model (e.g., Hertz model) for calculating Young's modulus, and document all fitting parameters [61].
- Solution: Establish and rigorously document a standard operating procedure (SOP) for your lab. This should include:
Guide 2: Ensuring Statistical Significance in Heterogeneous Biofilm Experiments
Problem: A study finds a weak or non-existent correlation between biofilm biomass and antibiotic susceptibility, raising questions about statistical power.
- Potential Cause: Small Sample Size (Low N). The study included too few bacterial strains or biofilm samples, leading to underpowered statistics that cannot reliably detect a true effect.
- Solution: Prioritize the use of "diverse and sufficiently large strain collections" [62]. Perform a power analysis before experimentation to determine the minimum sample size required to detect a meaningful effect. A systematic review highlighted that small sample sizes often lead to a lack of statistical support for observed correlations [62].
- Potential Cause: Inconsistent Biofilm Quantification Methods. Using different methods (e.g., Crystal Violet for total biomass vs. resazurin for metabolic activity) on the same set of samples can yield conflicting results, as they measure different aspects of the biofilm [62].
- Solution: Use multiple, complementary quantification methods in parallel and clearly report which method was used for each dataset. Be cautious when comparing results from studies that used different fundamental quantification techniques [62].
Problem: An experimental treatment appears to reduce biofilm biomass, but the results are not statistically significant.
- Potential Cause: High Variability in Biofilm Growth. Traditional liquid-culture biofilm models can exhibit high batch-to-batch variability, masking true treatment effects.
- Solution: Adopt more reproducible biofilm models. For example, the Modified Crone's Model (MCM), a semi-solid model, has been shown to display "reduced variability compared to liquid-culture systems" [63]. Ensure sufficient experimental replicates (n) are performed to account for biological variability.
Guide 3: Correlating Biofilm Structure with Function
Problem: It is difficult to link the physical structure of a biofilm (e.g., from AFM) to its functional outcome, such as antimicrobial tolerance.
- Potential Cause: Focusing Only on Biomass. Assuming that a larger biofilm is automatically a less susceptible one.
- Solution: Move beyond simple biomass measurements. The relationship between biofilm size and reduced susceptibility is "highly variable" and influenced by microbial species, strain-specific traits, and antibiotic type [62]. Investigate other structural parameters like extracellular matrix composition and spatial heterogeneity.
- Potential Cause: Lack of Objective Maturity Classification. Manually classifying biofilm maturity based on incubation time alone is subjective and prone to observer bias.
- Solution: Implement a standardized classification scheme. One study proposed a framework of 6 classes based on topographic characteristics from AFM. To avoid bias, you can use an open-access machine learning tool developed to classify staphylococcal biofilm maturity with accuracy comparable to human researchers [31].
Frequently Asked Questions (FAQs)
Q1: What are the most critical factors to control for reproducible AFM imaging of live biofilms? A1: The key factors are:
- Physiological Conditions: Image in liquid using a culture medium with controlled temperature and atmosphere to maintain cell viability [61].
- Scanning Mode: Use tapping mode to minimize sample damage [61].
- Sample Immobilization: Ensure the biofilm is firmly attached to a substrate to prevent detachment by the tip [61].
- Tip Quality: Always use a clean, undamaged tip [61].
Q2: Why do my biofilm susceptibility results differ from published literature? A2: This is a common challenge due to a "lack of standardized protocols" [62]. Differences can arise from:
- Biofilm Model: The choice of model (e.g., liquid vs. semi-solid) significantly impacts results. Susceptibility rankings can "differ substantially from traditional assays" based on the model used [63].
- Growth Conditions: Media, flow conditions, and incubation time can alter biofilm physiology.
- Susceptibility Metrics: Methods like Minimum Inhibitory Concentration (MIC) are not accurate for biofilms. Use biofilm-specific assays like Minimum Biofilm Eradication Concentration (MBEC) [62].
Q3: How can I objectively quantify the maturity of a biofilm beyond just incubation time? A3: You can use a classification scheme based on common topographic characteristics identified by AFM, such as the substrate coverage, bacterial cells, and extracellular matrix. To automate this and remove observer bias, a machine learning algorithm has been developed that can classify AFM images of staphylococcal biofilms into different maturity classes with high accuracy [31].
Q4: Can an electric field affect my biofilm measurements? A4: Yes, significantly. Research on 96-hour mature MRSA biofilms has shown that electric fields, such as those used in Electrochemical Impedance Spectroscopy (EIS), can cause a "destructive interaction" with the biofilm, leading to a "significative reduction of total biomass" in specific frequency ranges. This can alter the sample during measurement, questioning the reproducibility of electrical characterization techniques [64].
Q5: What is the relationship between the amount of biofilm (biomass) and its antibiotic susceptibility? A5: The relationship is complex and not deterministic. A systematic review found that the correlation is "highly variable". While some studies report that more biomass leads to less susceptibility, others show "weak or no such relationships". The data clearly indicate that 'bigger' biofilms are not by definition less susceptible [62]. Susceptibility is governed by multiple factors beyond size, including matrix composition and metabolic heterogeneity.
Quantitative Data on Biofilm Biomass & Susceptibility
The table below summarizes selected data from a systematic review investigating the correlation between biofilm biomass and antibiotic susceptibility, demonstrating the variability in findings. r² represents the goodness of fit, where a value of 1 indicates a perfect correlation and 0 indicates no correlation [62].
Table 1: Correlation Between Biofilm Biomass and Antibiotic Susceptibility
| Reference | Organism (No. of Isolates) | Biofilm Quantification Method | Antibiotic | Correlation (r²) Between Biomass and Susceptibility |
|---|---|---|---|---|
| Silva et al. [62] | S. aureus (n=23) | Crystal Violet (CV) | Tetracycline | 0.009 (Very Weak) |
| Silva et al. [62] | S. aureus (n=23) | Crystal Violet (CV) | Amikacin | 0.150 (Weak) |
| Silva et al. [62] | S. aureus (n=18) | Crystal Violet (CV) | Ciprofloxacin | 0.011 (Very Weak) |
| Wu et al. [62] | S. aureus (n=6) | Crystal Violet (CV) | Linezolid (6h) | 0.792 (Strong)* |
| Wu et al. [62] | S. aureus (n=6) | Resazurin Viability | Linezolid (6h) | 0.773 (Strong)* |
| Note: An asterisk () denotes a statistically significant correlation (p-value < 0.05). Adapted from [62].* |
Experimental Protocols
Protocol 1: AFM-Based Nanomechanical Profiling of Biofilms
Methodology for assessing biofilm stiffness and adhesion forces [61]:
- Sample Preparation: Grow biofilm on a sterile, rigid substrate (e.g., a PET slide or glass coverslip). For live cell imaging, mount the sample in a liquid cell containing an appropriate culture medium to maintain physiological conditions.
- Cantilever Selection and Calibration: Select a cantilever with an appropriate spring constant (typically soft, ~0.01-0.1 N/m for biofilms). Calibrate the spring constant of the cantilever using a thermal tuning method.
- AFM Setup: Engage the AFM tip in the liquid above the biofilm surface. Use tapping mode for topographical imaging to locate representative areas.
- Force Spectroscopy: Switch to force spectroscopy mode. Program the AFM to obtain force-distance curves at multiple predefined locations (e.g., a grid of 16x16 points) over the biofilm surface.
- Data Analysis: Analyze the retraction part of the force-distance curve to calculate adhesion forces. Analyze the approach curve using a contact mechanics model (e.g., Hertz model) to fit the data and extract Young's modulus (E), a measure of stiffness.
Protocol 2: Modified Crone's Model (MCM) for Semi-Solid Biofilm Culture
Methodology for a reproducible in vitro biofilm model that mimics tissue conditions [63]:
- Matrix Preparation: Prepare a soft-tissue-like matrix using agar-based material at a defined concentration.
- Bacterial Embedding: Suspend the bacterial strain in a suitable medium and mix it with the molten agar matrix at a temperature that does not harm the bacteria.
- Solidification: Dispense the mixture into the wells of a culture plate and allow it to solidify under sterile conditions.
- Incubation: Incubate the plate under conditions optimal for the bacterial strain (e.g., 37°C, aerobic) for the desired biofilm maturation period (e.g., 24-96 hours).
- Treatment and Analysis: Apply antimicrobial compounds directly onto the semi-solid surface. Biofilms can be harvested for analysis using methods like CV staining, resazurin assay, or colony-forming unit (CFU) counts.
Experimental Workflow Visualization
The diagram below illustrates a recommended workflow for conducting reproducible and statistically significant biofilm research, integrating AFM and susceptibility testing.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for Advanced Biofilm Research
| Item | Function/Brief Explanation | Key Context for Use |
|---|---|---|
| Crystal Violet (CV) Stain | Quantifies total adhered biofilm biomass (live/dead cells & matrix components). | A common, high-throughput method. Note it does not distinguish between live and dead cells [62]. |
| Resazurin Viability Stain | Measures the number of metabolically active cells in a biofilm. | Provides a different readout than CV; results from the two methods may not correlate [62]. |
| Atomic Force Microscope (AFM) | Provides high-resolution 3D topographical imaging and nanomechanical property measurement (stiffness, adhesion). | Ideal for characterizing heterogeneity at the nanoscale. Requires careful sample prep to avoid artifacts [61]. |
| Modified Crone's Model (MCM) | A semi-solid, agar-based biofilm model that mimics tissue-associated infections. | Offers more reproducible growth and in vivo-like morphology with reduced variability compared to liquid models [63]. |
| Machine Learning Classifier | An open-access software tool for automated classification of biofilm maturity from AFM images. | Reduces observer bias and standardizes maturity staging for staphylococcal biofilms [31]. |
| Confocal Laser Scanning Microscope (CLSM) | Enables high-resolution optical imaging of biofilms, often combined with fluorescent tags. | Used for validating biofilm structure and performing live/dead assays. Correlative use with AFM is powerful [64]. |
Establishing Internal Controls and Standards for Reliable Inter-Laboratory Data Comparison
Frequently Asked Questions (FAQs)
1. What are the most common sources of AFM artifacts when imaging biofilms? The most prevalent artifacts originate from tip-sample interactions, thermal drift, scanner nonlinearities, and improper feedback loop settings. Tip-related artifacts, including contamination and blunting, are exceptionally common and can lead to false topographical interpretations. [54]
2. How can we minimize thermal drift during long biofilm imaging sessions? Using closed-loop scanners with integrated position sensors provides real-time correction of positional inaccuracies. For systems without this hardware, allowing the instrument to thermally equilibrate before use and implementing post-acquisition drift correction algorithms can mitigate these effects. [54]
3. What is the best way to verify the accuracy of my AFM's lateral and vertical calibrations? Regular calibration using traceable standards is essential. For lateral (X-Y) calibration, use pitch gratings with known distances (e.g., 1-10 µm). For vertical (Z) calibration, use step height standards with precisely defined heights (e.g., 20-200 nm). Scan these standards under the same conditions used for your biofilm samples. [54]
4. Our lab studies live biofilms in liquid. How can we immobilize samples without inducing stress or artifacts? A non-perturbative protocol exists that avoids chemical or mechanical entrapment. Using indium-tin-oxide (ITO)-coated glass substrates can improve bacterial adhesion due to their hydrophobicity and smoothness, allowing for stable imaging of living bacteria in liquid culture medium without aggressive immobilization. [65]
5. Can machine learning assist in classifying biofilm maturity from AFM images? Yes. Deep learning algorithms can be trained to identify topographic characteristics associated with different biofilm maturity stages, such as the presence of individual cells, microcolonies, and extracellular matrix. One developed algorithm classifies staphylococcal biofilm images into six maturity classes with an accuracy comparable to human experts. [31]
Troubleshooting Guides
Guide 1: Addressing Tip-Related Artifacts
- Problem: Repeating, asymmetrical patterns or features that are much sharper than expected.
- Solution: This indicates a damaged or contaminated probe.
- Action 1: Image a known, sharp standard (e.g., TGT1 grating). If the features appear doubled or blurred, replace the probe.
- Action 2: Ensure using an appropriate cantilever for your mode (e.g., contact vs. tapping mode) and sample stiffness. Softer cantilevers (0.1 - 2 N/m) are often better for delicate biofilms. [54]
- Action 3: Implement automated probe conditioning systems if available. [3]
Guide 2: Correcting for Scanner Nonlinearities and Hysteresis
- Problem: Image distortion, particularly at the edges of scans, where features appear stretched or compressed.
- Solution:
- Action 1: Use a calibration grating with a regular, grid-like pattern to identify and correct for these distortions. Scan the grating over the same area size as your biofilm samples.
- Action 2: Prefer AFM systems with closed-loop scanners, which use sensors to actively correct the piezo position.
- Action 3: If using an open-loop system, restrict quantitative measurements to the center 60-70% of the scan area where nonlinearities are minimized. [54]
Guide 3: Optimizing Feedback Loop Parameters for Heterogeneous Samples
- Problem: "Parachuting" (loss of contact over steep features), streaks, or blurring on rough biofilm areas.
- Solution: Manually adjust the PID (Proportional, Integral, Derivative) gains.
- Action 1: On a flat region of the sample, increase the P-gain until the system begins to oscillate, then slightly reduce it.
- Action 2: Increase the I-gain to correct for long-term errors (offsets from the setpoint).
- Action 3: A small amount of D-gain can help dampen oscillations on sharp edges.
- Tip: Technologies like Bruker's ScanAsyst or PeakForce Tapping automate this optimization, which is highly beneficial for heterogeneous biofilms. [54]
Essential Calibration Standards and Quality Control
Regular calibration is non-negotiable for inter-laboratory comparison. The following standards should be incorporated into a routine quality control schedule.
Table 1: Essential AFM Calibration Standards for Biofilm Research
| Standard Type | Purpose (Parameter Measured) | Typical Specifications | Recommended Use Frequency |
|---|---|---|---|
| Step Height | Vertical (Z) Calibration, Linearity | Height: 20-200 nm (e.g., SiO2 steps on Si) | Before each new experiment series |
| Pitch Grating | Lateral (X-Y) Calibration | Pitch: 1-10 µm, Traceable to NIST | Weekly, or when changing objectives/scanners |
| Random Roughness | Resolution Verification, Tip Check | Ra ~ 100 nm | Monthly, or when tip damage is suspected |
| Honeycomb Pattern | Large-Area Stitching Accuracy | Millimeter-scale periodicity | For validating large-area AFM protocols [3] |
Experimental Protocol: Reliable Large-Area AFM of Early-Stage Biofilms
This protocol is adapted from methods used to study Pantoea sp. YR343 biofilm assembly, which revealed honeycomb patterns and flagellar interactions. [3] [66]
1. Sample Preparation (PFOTS-treated glass)
- Clean glass coverslips with oxygen plasma or piranha solution.
- Vapor-phase silanization: Place coverslips in a desiccator with a few drops of (Tridecafluoro-1,1,2,2-tetrahydrooctyl)trichlorosilane (PFOTS) under vacuum for 1 hour.
- Bake at 120°C for 30 minutes to complete the covalent bonding.
2. Bacterial Inoculation and Adhesion
- Grow Pantoea sp. YR343 (or your target strain) to mid-exponential phase.
- Dilute culture in fresh, appropriate growth medium to an OD660 of ~0.1.
- Incubate the PFOTS-treated coverslip in the bacterial suspension for a defined period (e.g., 30 minutes to 8 hours) at the desired temperature.
3. Sample Rinsing and Mounting
- Gently rinse the coverslip with sterile buffer (e.g., PBS or ultrapure water) to remove non-adherent planktonic cells.
- Critical Step: For imaging in air, allow the sample to dry naturally. For liquid imaging, mount the coverslip in the AFM liquid cell and submerge in an appropriate buffer.
4. Automated Large-Area AFM Imaging
- Use an AFM system capable of automated stage movement and image stitching.
- Mode: Tapping Mode in air or Quantitative Imaging (QI) Mode in liquid. [65]
- Set a grid of adjacent scan areas (e.g., 50x50 µm each) to cover a millimeter-scale region.
- Use a software-controlled stage to move between grid points automatically.
- Ensure a small overlap (e.g., 5-10%) between adjacent images for accurate stitching.
5. Data Processing and Machine Learning Analysis
- Stitching: Use software algorithms to merge the individual high-resolution images into a seamless large-area map. [3]
- Segmentation: Apply machine learning-based image segmentation to automatically detect individual cells, flagella, and other features.
- Quantification: Extract quantitative data such as cell count, surface coverage (confluency), cell orientation, and morphological parameters. [3]
Research Reagent Solutions
Table 2: Key Reagents and Materials for AFM Biofilm Studies
| Item | Function/Application | Example/Specification |
|---|---|---|
| PFOTS | Creates a hydrophobic, low-energy surface to promote specific bacterial adhesion patterns for consistent initial attachment studies. [3] | (Tridecafluoro-1,1,2,2-tetrahydrooctyl)trichlorosilane, >97% purity |
| ITO-coated Glass Slides | Provides a smooth, hydrophobic substrate that enhances bacterial adhesion for liquid-phase AFM without chemical immobilization, preserving native cell physiology. [65] | Surface roughness < 1 nm |
| Soft AFM Cantilevers | Minimizes applied force on delicate biofilm structures to prevent sample deformation and obtain accurate nanomechanical properties. | Spring constant: 0.1 - 2 N/m; Sharp tips (nominal radius < 10 nm) |
| Liquid Cell AFM Buffer | Maintains bacterial viability and native biofilm structure during in-liquid imaging. | e.g., 10 mM HEPES or PBS, pH 7.4 |
Workflow and Relationship Diagrams
AFM Artifact Correction Workflow
Data Reliability Framework
Conclusion
Effectively correcting AFM artifacts is not merely a technical necessity but a fundamental requirement for advancing our understanding of biofilms. By integrating the foundational knowledge of artifact origins with robust methodological protocols, intelligent troubleshooting, and rigorous validation through correlative microscopy, researchers can transform AFM from a qualitative imaging tool into a powerful quantitative platform. The future of reliable biofilm characterization lies in the continued adoption of automated large-area scanning, machine learning for real-time artifact detection and correction, and the development of standardized protocols. These advancements will directly enhance the precision of anti-biofilm therapeutic development, surface design for medical devices, and our fundamental knowledge of biofilm mechanics, ultimately leading to more effective clinical interventions against persistent biofilm-associated infections.
References
- 1. AFM Scanning Parameters and Image Artifacts [https://sassafras13.github.io/ImageArtifactsScanParams/]
- 2. Park Knowledge Portal [https://www.parksystems.com/en/learning-center/lc-detail.learning120.html]
- 3. Analysis of biofilm assembly by large area automated AFM [https://www.nature.com/articles/s41522-025-00704-y]
- 4. 4 Common Imaging Problems (and how to fix them) - NuNano [https://www.nunano.com/blog/2020/11/23/4-common-imaging-problems-and-how-to-fix-them]
- 5. AFM Artifacts - AFM FAQ - Frequently Asked Questions ... [https://www.fc.up.pt/pessoas/peter.eaton/artifacts/artifacts.html]
- 6. Quantification of bacterial adhesion forces using atomic ... [https://www.sciencedirect.com/science/article/abs/pii/S0167701299001372]
- 7. Sample Drift - AFM Artifacts - AFM FAQ [https://www.fc.up.pt/pessoas/peter.eaton/artifacts/sampledrift.html]
- 8. Tip Convolution Artefacts [http://gwyddion.net/documentation/user-guide-en/tip-convolution-artefacts.html]
- 9. Novel Combination of Atomic Force Microscopy and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2223268/]
- 10. Overcoming Challenges and Limitations Regarding the ... [https://www.mdpi.com/2079-6439/11/10/83]
- 11. Artificial intelligence-empowered imaging technologies for ... [https://www.sciencedirect.com/science/article/abs/pii/S1385894725084311]
- 12. Analysis of biofilm assembly by large area automated AFM [https://pmc.ncbi.nlm.nih.gov/articles/PMC12062311/]
- 13. Best practices and recommendations for accurate ... [https://www.sciencedirect.com/science/article/abs/pii/S0079670021000678]
- 14. A Brief Guide to AFM Probe Selection - Emma Benjaminson [https://sassafras13.github.io/ProbeSelection/]
- 15. Atomic force microscopy methodology and AFMech Suite ... [https://www.nature.com/articles/s41467-018-05902-1]
- 16. Contact Mode vs. Tapping Mode AFM Comparison [https://www.nanoandmore.com/contact-vs-tapping-mode-afm?srsltid=AfmBOoqrO4O_01cxFvPkhsDrS6OzC80dSJXw_09w7HWeEWn06-Mq_AUy]
- 17. Atomic Force Microscopy of Biofilms—Imaging, Interactions ... [https://www.intechopen.com/chapters/50739]
- 18. Soft matter analysis via atomic force microscopy (AFM) [https://www.sciencedirect.com/science/article/pii/S266652392300082X]
- 19. Atomic force microscopy-mediated mechanobiological ... - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC10842832/]
- 20. Absolute Quantitation of Bacterial Biofilm Adhesion and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2711294/]
- 21. Tips and Tricks for Frustrated AFM Users [https://www.afmworkshop.com/newsletter/frustrated-with-your-afm?srsltid=AfmBOoqs8in0juBJ1b6ps0ya2KZ_zRkZpTXismmbBO78DCiB3PIc9VKA]
- 22. Deep Learning for Live Cell Shape Detection and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9598706/]
- 23. Tissue section AFM: In situ ultrastructural imaging of native ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2877882/]
- 24. Tapping-mode atomic force microscopy in fluid of hydrated ... [https://www.sciencedirect.com/science/article/abs/pii/S0945053X01001743]
- 25. Frequently Asked Questions about Atomic Force Microscopy [https://www.afmhelp.com/index.php?option=com_content&view=article&id=49&Itemid=55]
- 26. Atomic Force Microscopy FAQs [https://www.bruker.com/en/products-and-solutions/microscopes/materials-afm/faq.html]
- 27. Quantitative Mechanical Property Mapping at the ... [https://www.bruker.com/en/products-and-solutions/microscopes/materials-afm/resource-library/an-128-quantitative-mechanical-property-mapping-at-the-nanoscale-with-peakforce-qnm.html]
- 28. High‐Speed Quantitative Nanomechanical Mapping by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12508714/]
- 29. Biofilm Cohesiveness Measurement Using a Novel Atomic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC1892862/]
- 30. New imaging approach transforms study of bacterial biofilms [https://www.ornl.gov/news/new-imaging-approach-transforms-study-bacterial-biofilms]
- 31. Machine learning assisted classification of staphylococcal ... [https://www.sciencedirect.com/science/article/pii/S2590207525000310]
- 32. Compensation Method for Correcting the Topography ... [https://www.mdpi.com/2075-1702/12/2/126]
- 33. AFM characterization of patterned sapphire substrate with ... [https://www.sciencedirect.com/science/article/abs/pii/S0169433217330106]
- 34. Atomic force microscopy for single molecule characterisation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6420408/]
- 35. Correction of the tip convolution effects in the imaging of ... [https://pubmed.ncbi.nlm.nih.gov/25201128/]
- 36. In-Situ Determination of the Mechanical Properties of Gliding ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0061663]
- 37. Compensating nonlinearities in high-speed atomic force ... [https://link.springer.com/article/10.1007/s12213-025-00194-3]
- 38. An ultra-wide scanner for large-area high-speed atomic ... [https://www.nature.com/articles/s41598-021-92365-y]
- 39. Mass transfer in heterogeneous biofilms - PubMed Central - NIH [https://pmc.ncbi.nlm.nih.gov/articles/PMC11416670/]
- 40. Chromatin Imaging with Time-Lapse Atomic Force ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4930652/]
- 41. Tips and Tricks for Frustrated AFM Users [https://www.afmworkshop.com/newsletter/frustrated-with-your-afm?srsltid=AfmBOoqPhNJFu5PeXAjCVe0LragOgO9tOn_EU8X7kunXkoElkGxAQOj3]
- 42. How to Optimize AFM scan parameters in 2min [https://www.nanoandmore.com/guide-to-optimizing-AFM-scan-parameters-settings?srsltid=AfmBOorcxcQbUJ6LPirw1U8f4l9i0YS5TPiF9JewZJnN-nB9UfnrfkK5]
- 43. Probing the reduction of adhesion forces between biofilms ... [https://www.sciencedirect.com/science/article/abs/pii/S0927776523005957]
- 44. DOE Scientists Develop Novel AFM Method to Transform ... [https://www.labcompare.com/617-News/620684-DOE-Scientists-Develop-Novel-AFM-Method-to-Transform-Biofilm-Research/?catid=61]
- 45. Indentation and Adhesive Probing of a Cell Membrane with AFM [https://pmc.ncbi.nlm.nih.gov/articles/PMC1366816/]
- 46. Atomic Force Microscopy - an overview [https://www.sciencedirect.com/topics/earth-and-planetary-sciences/atomic-force-microscopy]
- 47. Deciphering conformational dynamics in AFM data using ... [https://www.nature.com/articles/s42003-025-08365-5]
- 48. Applying the Atomic Force Microscopy Technique in ... [https://www.mdpi.com/2227-9059/12/9/2012]
- 49. Mapping of a Subgingival Dual-Species Biofilm Model ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2021.729720/full]
- 50. Errors and Mistakes to Avoid when Analyzing Raman Spectra [https://www.spectroscopyonline.com/view/errors-and-mistakes-to-avoid-when-analyzing-raman-spectra]
- 51. SEMTWIST Quantification of Biofilm Infection in Human ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12359142/]
- 52. Breaking the barrier: disruption of bacterial biofilms using ... [https://www.frontiersin.org/journals/cellular-and-infection-microbiology/articles/10.3389/fcimb.2025.1670237/full]
- 53. Optimization of fixation methods for observation of bacterial ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3181414/]
- 54. How To Tackle Artifacts In Atomic Force Microscopy Imaging [https://eureka.patsnap.com/report-how-to-tackle-artifacts-in-atomic-force-microscopy-imaging-solutions]
- 55. A guide for nanomechanical characterization of soft matter ... [https://www.sciencedirect.com/science/article/pii/S2666166725002151]
- 56. Evaluating large language model agents for automation of ... [https://www.nature.com/articles/s41467-025-64105-7]
- 57. How Microbes Use Force To Control Adhesion - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7253613/]
- 58. Advances in nanomechanical property mapping by atomic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12379807/]
- 59. Atomic Force Microscopy Application in Biological Research [https://pmc.ncbi.nlm.nih.gov/articles/PMC3700051/]
- 60. Atomic force microscopy study of morphological ... [https://www.sciencedirect.com/science/article/abs/pii/S0304399116301565]
- 61. applications of atomic force microscopy in disease [https://link.springer.com/article/10.1007/s12551-025-01306-w]
- 62. Is Increased Biofilm Formation Associated with Decreased ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12566419/]
- 63. A Semi-Solid In Vitro Biofilm Model for Evaluating ... [https://www.sciencedirect.com/science/article/pii/S2590207525000760]
- 64. On the effectiveness of electrical characterization of mature ... [https://www.nature.com/articles/s41598-025-93300-1]
- 65. Atomic Force Microsocopy: Key Unconventional Approach for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11603319/]
- 66. Analysis of biofilm assembly by large area automated AFM [https://pubmed.ncbi.nlm.nih.gov/40341406/]