Correlative AFM-Confocal Microscopy: A Multimodal Framework for Biofilm Nanomechanics and 3D Architecture
This article provides a comprehensive guide for researchers and drug development professionals on integrating Atomic Force Microscopy (AFM) and Confocal Laser Scanning Microscopy (CLSM) for advanced biofilm analysis.
Correlative AFM-Confocal Microscopy: A Multimodal Framework for Biofilm Nanomechanics and 3D Architecture
Abstract
This article provides a comprehensive guide for researchers and drug development professionals on integrating Atomic Force Microscopy (AFM) and Confocal Laser Scanning Microscopy (CLSM) for advanced biofilm analysis. It covers the foundational principles of AFM nanomechanics and CLSM imaging, detailed protocols for correlated methodology, strategies for troubleshooting common pitfalls, and a comparative validation against traditional techniques. By synthesizing insights from recent technological advancements, including automated large-area AFM and AI-driven analysis, this resource aims to empower the development of precision-guided therapeutic interventions against persistent biofilm-associated infections.
Understanding the Synergy: Core Principles of AFM Nanomechanics and Confocal Biofilm Imaging
Biofilms are sophisticated microbial communities encased in a self-produced matrix of extracellular polymeric substances (EPS), playing a critical role in up to 80% of persistent human infections [1] [2]. This structural complexity confers remarkable resilience against antimicrobial agents, with biofilm-embedded bacteria demonstrating up to 1,000-fold greater resistance compared to their planktonic counterparts [1]. The extracellular matrix functions as a formidable physical barrier, limiting antibiotic penetration while creating heterogeneous microenvironments that support metabolic dormancy and persister cell formation [1]. Understanding this intricate architecture is paramount for developing effective therapeutic strategies to combat biofilm-associated infections.
Advanced imaging technologies have revealed the spatial and chemical heterogeneity of biofilms, providing crucial insights into the structure-function relationships that underpin their recalcitrance. This guide examines how the correlation of atomic force microscopy (AFM) nanomechanics with confocal laser scanning microscopy (CLSM) structural data is transforming our understanding of biofilm resilience, enabling researchers to identify vulnerabilities and test novel intervention strategies.
Comparative Analysis of Biofilm Imaging Technologies
Technical Specifications and Capabilities
Table 1: Comparison of Key Biofilm Imaging Techniques
| Technique | Resolution | Imaging Environment | Key Measurable Parameters | Major Advantages | Inherent Limitations |
|---|---|---|---|---|---|
| Atomic Force Microscopy (AFM) | 2 nm (in-plane) [3] | Ambient air, liquid, vacuum [3] | Nanomechanical properties (Young's modulus, adhesion), 3D topography, molecular interactions [3] [4] [5] | Operates in physiological conditions; quantitative mechanical mapping; minimal sample preparation [3] [6] | Small imaging area (<100 μm) [7]; slow scanning speed; limited to surface properties [7] [3] |
| Confocal Laser Scanning Microscopy (CLSM) | ~200 nm [3] | Liquid, physiological conditions [8] [2] | 3D architecture, biofilm biovolume, live/dead cell distribution, spatial organization [8] [2] | Non-destructive optical sectioning; real-time monitoring of hydrated biofilms; molecular specificity with fluorescence [8] [2] | Requires fluorescent labeling; limited resolution compared to AFM; no mechanical property data [3] [2] |
| Scanning Electron Microscopy (SEM) | 10 nm [3] | High vacuum (conventional) [3] | Surface topology, cellular arrangement, extracellular matrix features [1] [9] | High-resolution surface imaging; large field of view [9] | Extensive sample preparation (dehydration, coating) causes artifacts [7] [9]; no living samples |
| Environmental SEM (ESEM) | 10-100 nm (varies) | Hydrated conditions possible [9] | Surface features of hydrated specimens | Direct visualization of hydrated biofilms [9] | Lower resolution than conventional SEM [9] |
Application-Based Performance
Table 2: Application Performance of AFM and CLSM in Biofilm Research
| Research Application | AFM Performance | CLSM Performance | Complementary Insights |
|---|---|---|---|
| Structural-Mechanical Correlation | Directly measures stiffness (Young's modulus) and adhesion forces of biofilm matrix [6] | Visualizes architectural features (porosity, thickness, microcolony formation) [6] | Links mechanical properties to specific structural features; identifies regional heterogeneity |
| Antimicrobial Efficacy Testing | Quantifies nanomechanical changes in response to treatment (e.g., stiffness reduction) [5] | Tracks penetration of fluorescent antimicrobials and resulting cell viability [2] | Correlates mechanical degradation with biological activity loss |
| Single-Cell Analysis | Measures mechanical properties of individual cells (e.g., cancer cells are softer than healthy cells) [5] | Resolves subcellular localization of fluorescent reporters and molecular constituents [10] | Connects cellular mechanics to functional states and phenotypic responses |
| Dynamic Process Monitoring | High-speed AFM tracks surface assembly and detachment events [3] | Time-lapse imaging captures 3D architectural development and population dynamics [8] | Reveals how mechanical and structural evolution are interconnected over time |
Experimental Protocols for Correlative AFM-Confocal Microscopy
Integrated Workflow for Structural and Mechanical Analysis
Diagram Title: Correlative AFM-Confocal Biofilm Analysis
Detailed Methodological Protocols
Sample Preparation Protocol
Biofilm Growth Conditions:
- Substrate Treatment: For bacterial adhesion studies, treat glass coverslips with PFOTS (perfluorooctyltrichlorosilane) to create a hydrophobic surface that promotes biofilm formation [7].
- Inoculation and Culture: Inoculate surfaces with bacterial suspension (e.g., Pantoea sp. YR343, Lactococcus lactis) in appropriate growth medium [7] [6]. For lab strains, use M17 broth with 0.5% glucose for Lactococcus lactis cultures [6].
- Incubation Parameters: Grow biofilms under static conditions at optimal growth temperature (e.g., 30°C for L. lactis, 37°C for human pathogens) for 24-48 hours to establish mature structures [6].
- Sample Stabilization: For AFM imaging in liquid, use dopamine-coated Petri dishes (4 mg/mL in PBS) to improve surface adhesion and maintain biofilm integrity during analysis [6].
Atomic Force Microscopy Protocol
Instrumentation and Calibration:
- Probe Selection: Use silicon or silicon nitride cantilevers with nominal spring constants of 0.01-0.1 N/m for soft biological samples [5] [6]. Calibrate each cantilever using thermal tuning method before measurement.
- Imaging Mode Selection: Employ force spectroscopy mode for nanomechanical mapping, collecting 2D arrays of force-distance curves (force volume technique) across the biofilm surface [5] [6].
- Measurement Parameters: Set approach/retract velocity between 0.5-2 μm/s with maximum indentation force of 0.5-2 nN to avoid sample damage while ensuring sufficient signal [6].
Data Acquisition and Analysis:
- Elasticity Measurement: Apply Hertz contact mechanics model to approach force-distance curves to calculate Young's modulus [5]. For thin samples on hard substrates, use modified models (Chen, Tu, or Cappella models) [5].
- Adhesion Forces: Analyze retraction curves to quantify adhesion forces from unbinding events [5] [6]. Use Johnson-Kendall-Roberts (JKR) or Derjaguin-Müller-Toporov (DMT) models for adhesive contact mechanics [5].
- Spatial Mapping: Generate 2D elasticity and adhesion maps with corresponding topography to visualize mechanical heterogeneity [6].
Confocal Laser Scanning Microscopy Protocol
Staining Procedures:
- Viability Staining: Use LIVE/DEAD BacLight bacterial viability kits with SYTO 9 and propidium iodide to distinguish live (green) from dead (red) cells within biofilms [2].
- Matrix Staining: Employ concanavalin A conjugated with Alexa Fluor dyes to label polysaccharide components of the EPS matrix [2].
- Specific Labeling: For engineered strains, utilize constitutive fluorescent protein expression (GFP, RFP) for species-specific identification in multi-species biofilms [8].
Imaging Parameters:
- Optical Configuration: Use 40x or 63x water-immersion objectives with numerical aperture >1.2 for optimal resolution in liquid samples [8].
- Z-stack Acquisition: Collect optical sections at 0.5-1 μm intervals through the entire biofilm thickness to reconstruct 3D architecture [8] [6].
- Multi-channel Detection: Configure separate detection channels for each fluorophore with appropriate emission filters to minimize cross-talk [8].
Image Analysis:
- Architectural Quantification: Use image analysis software (e.g., ImageJ, IMARIS) to calculate biovolume, porosity, surface area-to-volume ratio, and thickness [8] [6].
- Spatial Analysis: Implement spatial statistics to quantify clustering patterns and species distribution in multi-species biofilms [8].
Research Reagent Solutions for Biofilm Imaging
Table 3: Essential Research Reagents for Correlative AFM-Confocal Studies
| Reagent/Category | Specific Examples | Research Function | Application Notes |
|---|---|---|---|
| Fluorescent Labels | SYTO 9/propidium iodide, Concanavalin A-Alexa Fluor, constitutive GFP/RFP [8] [2] | Visualize viability, EPS components, and specific bacterial strains | Optimize concentration to minimize staining artifacts; confirm specificity with controls |
| Surface Modifiers | PFOTS, dopamine hydrochloride [7] [6] | Promote controlled biofilm adhesion to substrates | PFOTS creates hydrophobic surfaces; dopamine enables polymerization for stable anchoring |
| Culture Media | M17 broth with glucose, Brain Heart Infusion (BHI), semi-defined biofilm-promoting medium [8] [6] | Support reproducible biofilm growth under controlled conditions | Include selective antibiotics (spectinomycin, erythromycin) for plasmid maintenance in engineered strains |
| AFM Consumables | Silicon nitride cantilevers, colloidal probes [5] [6] | Nanomechanical property measurement and single-cell force spectroscopy | Calibrate spring constants for each cantilever batch; functionalize with specific ligands for adhesion studies |
Data Interpretation: Connecting Structure to Mechanics
Case Study: Lactococcus lactis Biofilm Analysis
A seminal study demonstrates the power of correlative AFM-CLSM analysis in elucidating how surface proteins influence biofilm mechanical properties [6]. CLSM revealed that piliated L. lactis strains formed heterogeneous, aerial biofilms with complex architectures, while non-piliated isogenic strains developed compact, uniform biofilms [6]. Corresponding AFM nanomechanical mapping showed striking differences: non-piliated strains formed stiffer biofilms (Young's modulus: 4-100 kPa) compared to piliated variants (0.04-0.1 kPa) [6]. This inverse relationship between structural complexity and mechanical stiffness highlights how surface appendages enable the formation of more porous, compliant biofilms that may facilitate nutrient diffusion and resistance to mechanical disruption.
Case Study: Pantoea sp. YR343 Spatial Organization
Large-area automated AFM combined with machine learning analysis revealed previously unrecognized spatial patterns during early biofilm formation [7]. High-resolution imaging over millimeter-scale areas identified a preferred cellular orientation among surface-attached cells, forming distinctive honeycomb patterns [7]. Detailed mapping of flagella interactions demonstrated that these appendages coordinate not only initial attachment but also subsequent biofilm assembly through intercellular connections [7]. This study exemplifies how advanced AFM methodologies can uncover organizational principles across multiple scales, bridging nanoscale interactions to emergent community architecture.
Emerging Technologies and Future Directions
Artificial Intelligence-Enhanced Biofilm Imaging
The integration of artificial intelligence with biofilm imaging represents a paradigm shift in analysis capabilities [10]. Deep learning algorithms enable automated segmentation of complex biofilm architectures, resolution enhancement beyond optical limits, and multimodal data fusion from complementary techniques [10]. Convolutional neural networks (CNNs) can precisely delineate biofilm boundaries in CLSM datasets, while generative adversarial networks reconstruct high-resolution structural information from lower-quality inputs [10]. These approaches facilitate high-throughput quantification of structural heterogeneity and dynamic processes that were previously intractable through manual analysis.
Large-Area AFM with Machine Learning
Traditional AFM has been limited by small imaging areas (<100 μm), restricting representativeness for heterogeneous biofilms [7]. Recent developments in automated large-area AFM address this limitation through coordinated sample positioning and automated image stitching, enabling high-resolution mapping over millimeter-scale areas [7]. When enhanced with machine learning algorithms for cell detection and classification, this approach captures spatial heterogeneity and rare architectural features that conventional methods routinely miss [7]. This technological advancement finally enables researchers to link nanoscale cellular interactions to the functional macroscale organization of biofilms.
Molecular Imaging Integration
Beyond structural and mechanical characterization, molecular imaging techniques provide complementary information about chemical composition and metabolic activity within biofilms [2]. Mass spectrometry imaging (MSI), Raman spectroscopy, and nuclear magnetic resonance (NMR) can spatially resolve the distribution of metabolites, quorum-sensing molecules, and antimicrobial compounds throughout the biofilm architecture [2]. The future of comprehensive biofilm analysis lies in multimodal correlation that simultaneously interrogates structural organization, mechanical properties, and molecular composition within the same biofilm sample.
Atomic Force Microscopy (AFM) has established itself as a dominant technique for characterizing mechanical properties at the nanoscale, transforming our understanding of material behavior in diverse fields from biology to materials science [11]. As a mechanical microscope, AFM operates by measuring the interaction force between a sharp probe and the sample surface, transducing this force into measurable cantilever deflection that can be converted back into quantitative force values [11]. This fundamental principle enables researchers to generate detailed nanomechanical maps—spatially resolved representations of mechanical parameters as a function of the tip's coordinates [11]. Unlike many other characterization techniques, AFM can perform these measurements under physiological conditions, making it particularly valuable for studying soft biological samples like living cells, bacteria, and biofilm matrices without extensive sample preparation that might alter their native properties [7] [12].
The application spectrum of AFM nanomechanical characterization is remarkably broad. In biological sciences, it has revealed that cancer cells exhibit different mechanical properties than healthy cells, opening avenues for prognostic applications [12]. In microbiology, AFM investigates how bacteria adapt to antibiotics, addressing the critical challenge of antimicrobial resistance [12]. Environmental scientists employ AFM to characterize micro- and nanoplastic particles in groundwater, analyzing their aggregation behavior and surface properties [13]. Recent technological advances have significantly enhanced AFM capabilities, with improvements in quantitative accuracy, spatial resolution, high-speed data acquisition, machine learning integration, and viscoelastic property mapping expanding the technique's utility across these diverse domains [11].
AFM Techniques for Nanomechanical Property Mapping
Classification of Nanomechanical Mapping Modes
AFM-based mechanical property measurements can be broadly categorized into two fundamental approaches: indentation and adhesion modes [11]. Indentation modes, the focus of this guide, involve applying a controlled deformation to the sample surface and analyzing the response to extract mechanical parameters. These methods can be further classified into three principal groups: force-distance curve-based methods, nanoscale rheology, and parametric methods [11]. Each approach offers distinct advantages and is suited to different sample types and research questions, with the choice of method depending on factors such as required spatial resolution, measurement speed, sample stiffness, and the specific mechanical properties of interest.
Table 1: Comparison of Major AFM Nanomechanical Mapping Techniques
| Technique | Fundamental Principle | Primary Measured Parameters | Typical Acquisition Speed | Best Suited Applications |
|---|---|---|---|---|
| Force Volume | Records force-distance curves at each pixel by modulating tip-sample distance [11] | Elastic modulus, adhesion force, deformation [11] [12] | Slow to moderate (traditional); up to 0.4 fps with advanced actuation [11] | Heterogeneous materials, single cells, polymer blends |
| Nano-DMA (Nanoscale Rheology) | Applies oscillatory signals to tip in contact and measures viscoelastic response [11] | Storage/loss moduli, loss tangent, complex modulus [11] | Moderate to fast | Viscoelastic materials, live cell dynamics, polymers |
| Parametric Methods | Drives cantilever at resonance and monitors oscillation parameter changes [11] | Elastic modulus, dissipation, stiffness [11] | Very fast | High-resolution mapping, dynamic processes |
| Multifrequency AFM | Excites and detects multiple cantilever frequencies simultaneously [13] | Elastic modulus, surface potential, composition [13] | Fast | Complex materials, microplastics, biological systems |
Technical Principles and Methodologies
Force Volume Mode represents the foundational approach for nanomechanical mapping, acquiring complete force-distance curves (FDCs) at each pixel of the sample surface [11]. These curves are generated by modulating the tip-sample distance while recording cantilever deflection, typically using triangular or sinusoidal waveforms [11]. The approach and retraction sections of FDCs provide complementary information: while approach curves primarily inform about elastic properties, retraction curves reveal adhesive interactions and inelastic processes [11]. Hysteresis between approach and retraction cycles indicates energy dissipation mechanisms, characteristic of viscoelastic materials like living cells [11]. The mechanical properties are extracted by fitting these experimental curves to contact mechanics models, with the Hertz model being most common for elastic properties, and Johnson-Kendall-Roberts (JKR) or Derjaguin-Müller-Toporov (DMT) models applied for adhesive contacts [12].
Nano-DMA techniques, inspired by macroscopic dynamic mechanical analysis, characterize viscoelastic properties by applying small oscillatory indentations to the sample while monitoring the time lag between the applied force and resulting deformation [11]. In practice, the tip is first approached to a predefined setpoint force (typically 1-20 nN) to establish a reference indentation depth of 100-500 nm, after which an oscillatory signal (10-50 nm amplitude) is applied while maintaining contact [11]. The viscoelastic properties are encoded in the phase shift between the driving excitation and the mechanical response, allowing calculation of storage and loss moduli that describe the elastic and viscous components of material behavior, respectively [11]. This method is particularly valuable for studying time-dependent mechanical properties in polymers and biological samples.
Parametric Methods, including bimodal AFM and contact resonance AFM, operate by driving the cantilever at its resonant frequency and monitoring changes in oscillation parameters (amplitude, phase, frequency) induced by tip-sample interactions [11]. These changes are then correlated with mechanical properties through analytical expressions or calibration procedures [11]. The significant advantage of parametric methods lies in their rapid acquisition speed, as they avoid the need for complete force-distance curves at each pixel. This enables high-speed mapping of mechanical properties, making them suitable for studying dynamic processes and reducing the risk of sample damage or tip wear during prolonged contact.
Experimental Protocols and Methodologies
Standardized Protocol for AFM Nanomechanical Characterization
Sample Preparation Requirements vary significantly depending on material type. For biological samples like biofilms, appropriate immobilization is crucial. Biofilms can be grown on AFM-compatible substrates such as glass coverslips, often treated with adhesion-promoting chemicals like PFOTS [7]. For protein samples, mica surfaces provide an atomically flat substrate; typical preparation involves adhering muscovite mica discs to support discs and air-drying before protein adsorption [14]. Maintaining physiological conditions is essential for living samples, requiring liquid imaging with appropriate buffers [7] [12].
Cantilever Selection and Calibration profoundly impact measurement accuracy. Soft cantilevers (spring constants 0.01-1 N/m) are preferred for biological samples to avoid excessive deformation or damage [12]. The precise determination of spring constant is typically performed using thermal tuning methods before measurements. Tip geometry must be considered when selecting contact mechanics models, with spherical tips often preferred for soft materials to minimize local stresses [12].
Measurement Parameters should be optimized for each sample type. For force volume mapping, maximum indentation force should be limited to prevent sample damage (typically 1-5 nN for cells and biofilms) [12]. Indentation depth should not exceed 10-20% of sample thickness to avoid substrate effects [12]. For nano-DMA measurements, oscillation amplitudes of 1-5 nm are typically used, with frequencies ranging from a few Hz to several hundred Hz depending on the specific viscoelastic relaxation processes under investigation [11].
Data Analysis Workflow begins with converting deflection vs. position data to force vs. separation curves. The contact point must be accurately identified, after which the indentation portion is fitted with appropriate contact mechanics models [12]. For heterogeneous samples, statistical analysis of parameter distributions across multiple locations and samples provides more meaningful characterization than single-point measurements.
Advanced Integrated Workflow for Correlated AFM-Confocal Biofilm Imaging
The integration of AFM nanomechanical data with confocal microscopy represents a powerful approach for comprehensive biofilm characterization, linking structural organization with mechanical properties.
Correlated Imaging Protocol begins with growing biofilms on optically transparent substrates suitable for both techniques. Initial confocal imaging identifies regions of interest based on architectural features while maintaining biofilm viability. Following confocal characterization, the same regions are located for AFM analysis without disturbing the sample. For mechanical mapping, force volume or nano-DMA modes are employed with appropriate parameters (soft cantilevers, minimal forces). Following AFM measurement, a second confocal scan verifies structural integrity and registers any changes.
Data Integration Challenges include coordinate system alignment between instruments and accounting for temporal evolution between sequential measurements. Recent advances address these limitations through specialized hardware that integrates both modalities and computational approaches using machine learning for image registration and data fusion [7] [10].
Large-Area AFM Implementation overcomes the traditional limitation of small scan sizes (<100 µm) that has restricted AFM's utility for studying millimeter-scale biofilm architectures [7]. Automated large-area AFM approaches systematically tile high-resolution images across extended regions, with machine learning algorithms seamlessly stitching adjacent scans and correcting for distortions [7]. This innovation enables investigation of spatial heterogeneity in mechanical properties across structurally complex biofilms, directly correlating local nanomechanical behavior with larger-scale organizational patterns observed in confocal microscopy [7].
Table 2: Research Reagent Solutions for AFM Nanomechanical Characterization
| Reagent/Equipment | Function/Application | Specification Guidelines |
|---|---|---|
| AFM Cantilevers | Force sensing and indentation | Spring constant: 0.01-1 N/m for soft materials; Tip geometry: spherical for homogeneous stress distribution [12] |
| Functionalized Tips | Specific molecular interactions | Chemical force microscopy: tips modified with specific functional groups or biomolecules [12] |
| Biofilm Substrates | Sample support and immobilization | PFOTS-treated glass coverslips; Freshly cleaved mica for protein immobilization [7] [14] |
| Imaging Buffers | Maintain physiological conditions | Phosphate-buffered saline (PBS) or appropriate growth media for living samples [7] [12] |
| Calibration Standards | Instrument verification | Reference samples with known mechanical properties (e.g., poly dimethyl siloxane) [12] |
Comparative Analysis with Alternative Techniques
Technique Capability Matrix
AFM nanomechanical characterization operates within a broader ecosystem of materials characterization techniques, each with distinct strengths and limitations. Confocal Laser Scanning Microscopy (CLSM) excels at non-invasive 3D structural imaging of hydrated biofilms with molecular specificity through fluorescent labeling but provides no direct mechanical information [7] [10]. Scanning Electron Microscopy (SEM) offers superior surface topographic resolution but requires sample dehydration and coating, potentially altering native mechanical properties and preventing liquid imaging [7]. Raman spectroscopy provides detailed chemical composition data but lacks spatial resolution for fine structural features and offers no mechanical property data [7] [10].
Table 3: Comparative Analysis of AFM vs. Alternative Characterization Techniques
| Technique | Spatial Resolution | Mechanical Property Data | Sample Requirements | Imaging Environment |
|---|---|---|---|---|
| AFM | ~1 nm lateral; ~0.1 nm vertical [7] | Comprehensive: elasticity, adhesion, viscoelasticity [11] [12] | Minimal preparation; can be native state | Ambient, liquid, or controlled gas [7] [12] |
| Confocal Microscopy | ~200 nm lateral; ~500 nm axial [10] | None | Fluorescent labeling often required | Primarily liquid for biological samples |
| Scanning Electron Microscopy (SEM) | ~1 nm lateral [7] | None | Dehydration, conductive coating | High vacuum typically required [7] |
| Raman Spectroscopy | ~300-500 nm lateral [10] | Indirect inference only | Minimal for surface-enhanced approaches | Ambient or liquid possible |
AFM's unique capability to provide quantitative mechanical property data with high spatial resolution under physiological conditions represents its most significant advantage over alternative techniques. This enables researchers to establish direct structure-function relationships in native environments, particularly valuable for dynamic biological systems like biofilms [7] [12].
Performance Benchmarking in Biofilm Research
In the specific context of biofilm research, AFM demonstrates particular advantages for investigating early attachment events and surface interactions at the single-cell level. High-resolution AFM imaging has revealed intricate details of bacterial flagella and their coordination during surface attachment, showing flagellar structures measuring approximately 20-50 nm in height and extending tens of micrometers across surfaces [7]. These observations provide mechanical insights into initial biofilm formation that are inaccessible to optical techniques.
For investigating the structural role of extracellular polymeric substances (EPS) in biofilm mechanics, AFM has identified distinctive mechanical signatures associated with matrix components, enabling correlation of local composition with mechanical function [7]. When integrated with confocal microscopy through correlated imaging workflows, AFM mechanical data can be directly linked with spatial organization and compositional heterogeneity observed over larger areas, creating comprehensive models of biofilm behavior [7] [10].
The application of machine learning and artificial intelligence is transforming AFM capabilities in biofilm research, automating processes such as image segmentation, cell detection, and classification [7]. These advances enable high-throughput analysis of biofilm mechanical properties across statistically relevant areas, overcoming previous limitations from small sample sizes and operator-dependent variability [7] [10].
Applications in Biofilm Research and Beyond
Case Study: Nanomechanical Mapping of Bacterial Biofilms
Recent research employing large-area automated AFM has revealed remarkable spatial heterogeneity in the mechanical properties of developing biofilms. Studies of Pantoea sp. YR343 biofilms demonstrated distinct organizational patterns during early formation, with cells orienting in specific directions to form characteristic honeycomb structures [7]. AFM nanomechanical mapping correlated these architectural arrangements with local variations in mechanical properties, providing insights into how structural organization influences biofilm integrity and function.
The detailed characterization of flagellar interactions through AFM imaging suggests that flagellar coordination plays a role in biofilm assembly beyond initial attachment [7]. These mechanostructural investigations at the single-cell level provide foundational knowledge for developing strategies to control biofilm formation in industrial and medical contexts.
Environmental and Biomedical Applications
In environmental science, AFM nanomechanical characterization has proven valuable for investigating micro- and nanoplastic (MNP) particles in groundwater systems [13]. Multifrequency AFM techniques distinguish pristine and aged MNPs through differences in elastic modulus while simultaneously providing detailed morphological and roughness data [13]. These investigations reveal that environmental MNPs form aggregates with surface roughness one to two orders of magnitude higher than laboratory-aged particles, suggesting significantly different adsorption capacities for environmental pollutants [13].
In biomedical applications, AFM has revealed that cancer cells exhibit different mechanical properties than their healthy counterparts, with malignant cells typically showing decreased stiffness [12]. Although more sophisticated investigations are required for reliable prognostic applications, these mechanical differences offer potential diagnostic avenues. Similarly, AFM investigations of bacterial response to antibiotics provide insights into mechanisms of antimicrobial resistance, while studies of virus capsids have correlated increased stiffness with reduced infectivity, informing new antiviral strategies [12].
The field of AFM nanomechanical characterization continues to evolve rapidly, with several emerging trends shaping its future development. The integration of artificial intelligence and machine learning is enhancing data acquisition, analysis, and interpretation across multiple domains [11] [7] [10]. In biofilm research specifically, AI-driven approaches are enabling automated segmentation, classification, and analysis of complex structural and mechanical data, facilitating high-throughput investigation of biofilm heterogeneity and response to environmental perturbations [7] [10].
Advances in computational methods are bridging the resolution gap between AFM topographic imaging and atomic-scale structural models. Tools like AFMfit and NMFF-AFM employ flexible fitting procedures to deform atomic models to match multiple AFM observations, generating conformational ensembles that describe experimental data with atomistic precision [15] [16]. Similarly, the ProFusion framework enables 3D reconstruction of protein complex structures from multi-view AFM images using deep learning models trained on synthetic AFM data [14]. These approaches are extending AFM's utility from structural characterization to understanding dynamic molecular processes.
For correlated AFM-confocal studies of biofilms, future developments will likely focus on real-time integrated imaging systems that eliminate the temporal gap between measurements. Combined with machine learning algorithms for data fusion and analysis, these advances will enable comprehensive four-dimensional characterization of biofilm development, correlating structural, chemical, and mechanical evolution across spatial scales from single molecules to multicellular communities.
In conclusion, AFM nanomechanical characterization provides unique capabilities for investigating material properties across diverse research domains. Its particular strength lies in correlating mechanical behavior with structural features at the nanoscale under physiologically relevant conditions. When integrated with complementary techniques like confocal microscopy, AFM enables researchers to establish comprehensive structure-function relationships in complex biological systems, offering powerful insights for biomedical, environmental, and materials applications.
Confocal Laser Scanning Microscopy (CLSM) is a high-resolution fluorescence imaging technique that has become a cornerstone tool in biomedical research for visualizing the three-dimensional architecture of biological samples and assessing cell viability. Its core principle lies in the use of spatial filtering to achieve optical sectioning, providing a significant advantage over conventional widefield fluorescence microscopy [17]. In CLSM, a laser of specific excitation wavelength is scanned across the specimen, and the emitted fluorescent signal is detected only from the plane currently in focus, with out-of-focus light excluded by a pinhole aperture placed in front of the detector [17]. This mechanism enables the acquisition of sharp images from a single focal plane within fluorescently labeled specimens.
The capability to reject out-of-focus light allows researchers to construct detailed three-dimensional images by sequentially scanning and stacking optical sections (z-stacks) at different depths [17]. This is particularly valuable for studying complex structures such as biofilms, tissue models, and cellular spheroids, where understanding the 3D organization is critical. Furthermore, the digital nature of CLSM images facilitates advanced image processing and quantitative analysis, making it an indispensable tool for researchers, scientists, and drug development professionals who require precise morphological and viability data from within 3D microenvironments [17].
Figure 1: Optical sectioning in CLSM. A pinhole blocks out-of-focus light, collecting signal only from the focal plane.
Visualizing 3D Architecture and Quantifying Viability
Resolving Three-Dimensional Microenvironments
CLSM's optical sectioning capability enables the non-invasive exploration of 3D biological structures at high resolution. Under optimal conditions, CLSM can achieve a spatial resolution of approximately 0.2 × 0.2 × 0.8 micrometers (X, Y, Z), supporting detailed analyses of subcellular structures [17]. This has proven invaluable for characterizing the architecture of hepatocyte spheroids (HCS), which develop a multilayered structure with site-dependent cell viability, including highly viable cell layers, low active cell layers, and necrotic cores [18]. Similarly, in biofilm research, CLSM allows for in-depth analysis of the extracellular polymeric matrix without killing or damaging the biological structure, providing insights into biofilm composition, structure, and metabolism [19].
The technique has also been applied to study the distribution of fluorescent proteins (FPs) in coral tissues at the cellular level, revealing species-specific patterns. For instance, in Stylophora corals, green fluorescent proteins (GFPs) were concentrated in the intermesenterial muscle bands of the polyp, while cyan fluorescent proteins (CFPs) were located at the tips of the tentacles [20]. Such high-resolution visualization of pigment distribution helps elucidate photobiological adaptations and stress responses.
Quantitative Viability Assessment
A key application of CLSM is the quantitative assessment of cell viability within 3D structures. This is typically achieved using viability indicators and fluorescent stains that distinguish live from dead cells based on membrane integrity or metabolic activity [19]. For example, in hepatocyte spheroids, cell staining has been used as an indicator of cell viability as determined by the integrity of the cell wall membrane [18]. However, the large diameter and dense multilayered structure of thick specimens can result in uneven penetration of staining reagents, causing intrinsic fluorescence intensity errors in imaging and affecting the accurate measurement of cell viability at the center [18]. This limitation necessitates careful interpretation of viability data from deep within 3D samples.
CLSM has been benchmarked against other viability assessment methods. In a study quantifying site-dependent cell viability in hepatocyte spheroids, CLSM results were compared with those obtained from dynamic optical coherence tomography (D-OCT), a non-invasive, label-free method that detects cellular activity based on intrinsic motion patterns [18]. The study found that cells in C3A hepatocyte spheroids were active mainly in the range of 8 to 13 Hz when detected by D-OCT, and the D-OCT results showed a high degree of correspondence with CLSM data, validating both approaches for viability assessment [18].
Comparative Analysis: CLSM vs. Alternative Imaging Modalities
The selection of an appropriate imaging technique depends on the specific research requirements, including resolution needs, sample thickness, viability constraints, and the necessity for live or dynamic imaging. The table below provides a structured comparison of CLSM with other prominent bioimaging techniques.
Table 1: Technical comparison of CLSM with other bioimaging techniques
| Technique | Resolution | Imaging Depth | Viability Assessment | Key Advantages | Primary Limitations |
|---|---|---|---|---|---|
| CLSM | ~0.2 × 0.2 × 0.8 µm (X, Y, Z) [17] | ~100-150 µm [17] | Fluorescent viability stains (e.g., membrane integrity dyes) [19] [18] | Optical sectioning, 3D reconstruction, reduced out-of-focus blur [17] | Photobleaching, phototoxicity, limited depth [17] [21] |
| Multiphoton Microscopy | Similar to CLSM [17] | Up to 2-3 times deeper than CLSM [17] | Same fluorescent stains as CLSM | Deeper tissue penetration, reduced photobleaching outside focal plane [17] | Higher cost, complex laser systems [17] |
| Light Sheet Fluorescence Microscopy (LSFM) | Varies with objective NA | Several mm (with clearing) [21] | Same fluorescent stains as CLSM | Very low phototoxicity, fast volumetric imaging [21] | Lower resolution than CLSM, requires specialized sample mounting [21] |
| Dynamic Optical Coherence Tomography (D-OCT) | Axial: 1-10 µm, Lateral: 1-10 µm [18] | 1-3 mm [18] | Label-free, based on cellular motility frequencies [18] | Non-destructive, label-free, deep penetration [18] | No molecular specificity, indirect viability measure [18] |
| Widefield Fluorescence Microscopy | Diffraction-limited | Limited by scattering | Fluorescent viability stains | Simple, fast, cost-effective [1] | Out-of-focus blur, unsuitable for thick samples [17] [21] |
| Scanning Electron Microscopy (SEM) | Nanometer scale | Surface imaging only | Not applicable for live cells | Ultra-high resolution surface details [1] | Requires fixation, no live imaging [17] |
Table 2: Performance comparison for specific research applications
| Application | CLSM Performance | Competitive Technique Performance | Key Differentiating Factors |
|---|---|---|---|
| Biofilm Structure Analysis | High-resolution 3D visualization of matrix and bacterial colonies [19] | SEM: Higher surface resolution but no live imaging [1] | CLSM enables live imaging of hydrated biofilms; SEM provides superior surface detail but requires fixation [19] [1] |
| Cell Viability in 3D Spheroids | Quantitative with fluorescent stains; limited by dye penetration in dense spheroids [18] | D-OCT: Label-free viability assessment based on cellular motility [18] | D-OCT enables long-term non-invasive monitoring; CLSM provides molecular specificity but risks phototoxicity [18] |
| Deep Tissue Imaging | Limited to ~100-150 µm due to light scattering [17] | Multiphoton: 2-3 times deeper penetration than CLSM [17] | Multiphoton's near-infrared excitation scatters less, enabling deeper imaging in scattering tissues [17] |
| Long-term Live Cell Imaging | Phototoxicity limits duration [21] | LSFM: Significantly reduced phototoxicity [21] | LSFM illuminates only the detected plane, enabling longer studies with minimal damage [21] |
| Correlative Nanomechanics + Fluorescence | Compatible with AFM integration [22] | Super-resolution SIM + AFM: Higher optical resolution [22] | SR-SIM provides ~2x better resolution than CLSM but requires more complex reconstruction [22] |
Experimental Protocols for Key Applications
Protocol: Viability Assessment in 3D Spheroids
The following protocol details the methodology for evaluating cell viability in 3D hepatocyte spheroids using CLSM, as referenced in [18]:
- Spheroid Generation: Prepare hepatocyte spheroids using a liquid overlay technique on concave-bottomed culture wells with low adhesion. Inoculate C3A hepatocyte suspensions at concentrations of 10⁵ cells/ml with varying volumes (20-100 µl) to control spheroid size.
- Culture Conditions: Maintain spheroids in specialized culture medium at 37°C and 5% CO₂, replacing medium every 48 hours.
- Viability Staining: Apply appropriate fluorescent viability stains. Common approaches include:
- Membrane integrity dyes: Such as propidium iodide (dead cells) combined with calcein-AM (live cells).
- Metabolic indicators: That reflect cellular activity.
- CLSM Imaging:
- Use objectives with appropriate numerical aperture (e.g., 40×/0.60 or 63×/1.4 oil immersion).
- Set laser wavelength to match fluorophore excitation (e.g., 552 nm).
- Acquire z-stacks through entire spheroid with optimal step size (e.g., 1-5 µm).
- Maintain minimal laser power to reduce phototoxicity.
- Image Analysis:
- Reconstruct 3D volumes from z-stacks.
- Segment viable and necrotic regions based on fluorescence signals.
- Quantify viable cell volume ratio relative to total spheroid volume.
Protocol: CLSM and AFM Correlative Imaging
This protocol outlines the procedure for correlative CLSM and Atomic Force Microscopy (AFM) imaging, enabling simultaneous topological, nanomechanical, and fluorescence characterization [22]:
- Sample Preparation:
- Culture cells expressing fluorescently tagged proteins (e.g., EGFP-MCT1) on glass coverslips compatible with both techniques.
- Alternatively, prepare biofilm samples with appropriate fluorescent labeling of matrix components.
- Instrument Setup:
- Mount AFM on inverted CLSM microscope.
- Select cantilevers appropriate for biological samples (e.g., nominal resonance frequency of 90 kHz in air, spring constant of 0.3 Nm⁻¹).
- Align AFM laser spot on cantilever using CCD camera.
- Simultaneous Imaging:
- Engage AFM in force spectroscopy-based mode (e.g., Quantitative Imaging mode).
- Set Z-cantilever velocity and maximum Z-length (e.g., 250 µm/s at max Z-length of 1.5 µm).
- Acquire CLSM images simultaneously using appropriate laser wavelengths and detection settings.
- For fluorescence acquisition, use high-aperture objectives (e.g., 100× Oil, NA 1.49).
- Data Correlation:
- Overlay AFM topological/mechanical maps with CLSM fluorescence images.
- Correlate nanomechanical properties with fluorescence localization patterns.
Figure 2: Workflow for correlative CLSM and AFM imaging, combining fluorescence with nanomechanical data.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Essential research reagents and materials for CLSM experiments
| Reagent/Material | Function | Example Applications | Considerations |
|---|---|---|---|
| Fluorescent Viability Stains | Distinguish live/dead cells based on membrane integrity or metabolic activity | Viability assessment in 3D spheroids, biofilms [18] | Penetration efficiency in dense structures varies; potential phototoxicity |
| Genetically Encoded Fluorescent Proteins (e.g., GFP, YFP) | Label specific proteins or cell types; reporter gene expression | Visualization of protein localization, gene expression patterns [17] [22] | Enables long-term studies without external staining; may require transfection/transduction |
| Immunofluorescence Labels | Target-specific staining of cellular components | Identification of specific cell types, extracellular matrix components [17] | Requires sample fixation; enables multiplexing with multiple antibodies |
| Refractive Index Matching Solutions | Reduce light scattering in thick samples | Deep tissue imaging, improved penetration in 3D samples [21] | May not be compatible with live cell imaging; ongoing development of biocompatible options |
| Specialized CLSM Objectives | High-resolution light collection | All CLSM applications | High numerical aperture (e.g., 1.4) objectives critical for optimal resolution [23] |
| Photostabilizing Reagents | Reduce photobleaching during extended imaging | Long-term time-lapse experiments | Enables longer imaging sessions but may have biological effects |
Integration with Atomic Force Microscopy for Correlative Nanomechanics
The integration of CLSM with Atomic Force Microscopy (AFM) creates a powerful correlative imaging platform that combines nanomechanical characterization with biochemical specificity. This combination is particularly valuable for biofilm research, where understanding the relationship between mechanical properties and 3D structure is essential [4] [22]. AFM provides quantitative measurements of mechanical properties including stiffness and viscosity with sub-cellular sensitivity, while CLSM simultaneously maps the 3D distribution of fluorescently labeled components [4].
Technical implementations of combined CLSM-AFM platforms have demonstrated successful simultaneous operation, though careful system design is required to minimize interference [22]. For instance, the fluorescence excitation light must be controlled to prevent perturbation of the AFM cantilever operation [22]. Similarly, noise transfer from the optical microscope to the AFM must be mitigated to maintain measurement integrity. Despite these challenges, the correlative approach provides a mechanistic understanding of biological processes governing the unique functions of tissues and microbial communities [4].
This integrated methodology is particularly relevant for investigating how the extracellular polymeric matrix contributes to the mechanical stability of biofilms, or how cellular mechanical properties change during differentiation in 3D spheroids. The combination of AFM-based tissue mechanobiology with CLSM visualization enables researchers to correlate changes in mechanical properties with specific biochemical or structural alterations occurring during development, physiological adaptation, or disease progression [4].
Biofilms are complex, three-dimensional microbial communities embedded in a self-produced extracellular polymeric substance (EPS) matrix. This architecture is fundamental to their resilience against antimicrobial agents and host immune responses, posing significant challenges in medical and industrial contexts [24] [1]. Understanding the intricate structure-function relationships within biofilms requires imaging techniques that can capture both their detailed nanoscale surface properties and their broader 3D organization. While numerous microscopy methods are employed in biofilm research, Atomic Force Microscopy (AFM) and Confocal Laser Scanning Microscopy (CLSM) represent two powerful but fundamentally different approaches. Independently, each technique provides a valuable yet incomplete picture; AFM excels in surface topography and nanomechanics, whereas CLSM is unparalleled for 3D architectural visualization within hydrated, living samples. This guide objectively compares the performance of AFM and CLSM and demonstrates, through experimental data and protocols, why their correlative application is imperative for a holistic understanding of biofilm dynamics, mechanics, and response to external stimuli.
Technique Comparison: Strengths and Limitations
The following table summarizes the core capabilities of AFM and CLSM, highlighting their complementary nature.
Table 1: Core Capabilities of AFM and CLSM in Biofilm Research
| Feature | Atomic Force Microscopy (AFM) | Confocal Laser Scanning Microscopy (CLSM) |
|---|---|---|
| Resolution | Sub-nanometer vertical; molecular-scale lateral [7] [25] | Diffraction-limited (~200 nm lateral, ~500 nm axial) [26] |
| Key Outputs | 3D topography, nanomechanical properties (elasticity, adhesion), molecular interaction forces [24] [5] [25] | 3D architecture, biovolume, thickness, spatial distribution of labeled components [24] [27] |
| Sample Environment | Can operate in liquid under physiological conditions [28] [25] | Ideal for hydrated, living samples; requires immersion objective [29] [27] |
| Sample Preparation | Minimal; often requires immobilization but no staining [25] | Requires fluorescence labeling (e.g., dyes, tags) which may alter biofilm [24] |
| Primary Limitation | Small scan area (<150×150 µm); surface probe only; slow imaging; potential sample damage [7] [25] | Limited resolution; photobleaching; signal interference from intrinsic biofilm fluorescence [24] |
| Key Application | Quantifying adhesion forces, single-cell stiffness, EPS viscoelasticity [24] [5] | Visualizing live/dead cell stratification, EPS composition, and real-time structural dynamics [24] [27] |
The Synergy of Correlative AFM-CLSM
The limitations of each technique are directly addressed by the strengths of the other. AFM's small field of view makes it difficult to locate representative regions of interest within a heterogeneous biofilm and to contextualize nanoscale measurements within the broader biofilm architecture [7]. Conversely, while CLSM effectively maps the 3D structure, it lacks the resolution and capability to quantify the mechanical properties of the EPS matrix and individual cells, which are critical to biofilm stability and function [24] [28]. By combining them, researchers can pinpoint specific locations with CLSM and then use AFM to perform detailed topographical and nanomechanical analyses at those sites.
This synergy is powerfully illustrated in a 2018 study that combined Optical Coherence Tomography (OCT), a meso-scale imaging technique, with AFM to investigate oral biofilms. The research revealed that increasing sucrose concentration decreased the biofilm's Young's modulus (a measure of stiffness) while increasing cantilever adhesion, changes linked to variations in EPS content visualized via OCT [28]. This study exemplifies the "correlative imperative"—the structural insights from imaging informed the interpretation of the mechanical data, creating a comprehensive structure-property relationship unattainable with either technique alone.
Table 2: Quantitative Parameters Accessible via AFM and CLSM
| Parameter | Technique | Significance in Biofilm Research |
|---|---|---|
| Young's Modulus | AFM [28] [5] | Indicates biofilm stiffness; influences antimicrobial penetration and mechanical stability [24]. |
| Adhesion Forces | AFM [28] [25] | Quantifies cell-surface and cell-cell interactions, crucial for initial attachment and cohesion [24]. |
| Biovolume | CLSM [24] [27] | Measures total biomass, useful for assessing biofilm growth and treatment efficacy [27]. |
| Biofilm Thickness | CLSM [29] [27] | Determines the vertical dimension of the biofilm, related to nutrient gradients and metabolic activity. |
| Surface Roughness | AFM [30] | Influenced by microcolony formation; a parameter for quantitative morphological analysis [24]. |
| Substratum Coverage | CLSM [27] | Percentage of surface area covered by biofilm; indicates adhesion and spreading. |
Experimental Protocols for Correlative Imaging
Implementing a correlative AFM-CLSM workflow requires careful planning to maintain sample integrity between measurements. Below is a generalized protocol for a sequential analysis, where the same sample is first imaged with CLSM and then with AFM.
Correlative Workflow Protocol
Step 1: Sample Preparation and Immobilization
- Substrate: Use a thin, sterile glass coverslip or a specialized AFM dish as the growth substrate. The material must be compatible with high-resolution optics for CLSM and flat enough for stable AFM scanning.
- Biofilm Cultivation: Grow the biofilm on the substrate under controlled conditions relevant to your research question (e.g., flow or static culture) [29].
- Staining (for CLSM): After cultivation, gently rinse the biofilm with an appropriate buffer (e.g., PBS) to remove non-adherent cells. Apply fluorescent stains, such as SYTO 9 for total cells and propidium iodide for dead cells, or conjugated lectins for specific EPS components [24] [27]. Incubate in the dark, then rinse again to remove excess dye.
Step 2: Confocal Laser Scanning Microscopy
- Mounting: Place the stained, hydrated biofilm sample on the CLSM stage. Ensure it is fully submerged in buffer to prevent dehydration.
- Imaging: Use appropriate laser lines and emission filters for your chosen fluorophores. Acquire z-stacks through the entire biofilm thickness with a step size of 0.5-1.0 µm to reconstruct the 3D architecture.
- Data Analysis: Use image analysis software (e.g., ImageJ, IMARIS, or instrument-specific software) to calculate parameters like biovolume, thickness, and substratum coverage [27]. Crucially, note the X-Y-Z coordinates of specific regions of interest (ROIs), such as microcolony centers, water channels, or areas with varying fluorescence intensity.
Step 3: Atomic Force Microscopy
- Sample Transfer: Carefully transfer the sample from the CLSM stage to the AFM, keeping it hydrated throughout the process.
- Navigation: Use the optical microscope integrated with the AFM to navigate to the previously mapped ROIs using the recorded coordinates and distinctive morphological features.
- AFM Imaging & Force Spectroscopy:
- Topography Imaging: Perform contact or tapping mode scans in liquid to obtain high-resolution topographical images of the selected ROIs.
- Force Volume Imaging: Acquire 2D arrays of force-distance curves over the ROI. This generates simultaneous maps of topography and mechanical properties like Young's modulus and adhesion [28] [5].
- Probe Selection: Use sharp tips (nominal spring constant ~0.1 N/m) for high-resolution imaging and spherical tips (or colloidal probes, ~0.3-0.4 N/m) for more quantitative, reproducible nanomechanical mapping [28].
Diagram 1: Correlative AFM-CLSM experimental workflow for biofilm analysis.
Key Research Reagent Solutions
Table 3: Essential Materials for Correlative AFM-CLSM Biofilm Experiments
| Item | Function/Application | Brief Explanation |
|---|---|---|
| Glass Bottom Dishes/Petri Dishes | Sample Substrate | Provides a optically clear and flat surface essential for high-resolution CLSM and stable AFM scanning. |
| Fluorescent Stains (e.g., SYTO 9, Propidium Iodide, Conjugated Lectins) | Biomass and Component Labeling | Allows visualization of different biofilm components (live/dead cells, specific polysaccharides) in CLSM. |
| Phosphate Buffered Saline (PBS) | Buffer | Maintains physiological conditions (pH, osmolarity) during rinsing and imaging to preserve native biofilm structure. |
| AFM Cantilevers | Probing the Sample | The choice of tip (sharp vs. colloidal) determines the resolution and type of nanomechanical data acquired. |
| Calibration Grids (e.g., TGZ1-TGZ3) | AFM Instrument Calibration | Verifies the scanner's accuracy in X, Y, and Z dimensions, ensuring reliable and quantifiable measurements. |
Advanced Applications and Future Directions
The correlative approach is being supercharged by technological advancements. A landmark 2025 study introduced an automated large-area AFM approach capable of scanning millimeter-scale areas, a significant leap from the traditional sub-150 µm range [7]. This method, aided by machine learning for image stitching and cell classification, revealed a preferred cellular orientation and a distinctive honeycomb pattern in Pantoea sp. YR343 biofilms, features previously obscured by the limited field of view. When correlated with CLSM, such large-area AFM datasets can directly link nanoscale cellular arrangements and flagellar interactions observed by AFM to the overall community architecture visualized by CLSM.
Future directions point towards even deeper integration. Machine learning and AI are transforming AFM by automating routine tasks, optimizing scanning, and enhancing data analysis, making the correlation of large, information-rich datasets from both techniques more efficient [7]. Furthermore, the development of microfluidic flow cells that are compatible with both AFM and CLSM allows for the real-time observation of biofilm development, structural changes, and mechanical evolution under controlled hydrodynamic and chemical conditions [29]. This enables researchers to not only correlate structure and mechanics but also to observe the dynamic interplay between them in response to treatments, such as the introduction of antimicrobial agents or mechanical stress.
Diagram 2: How AFM and CLSM illuminate different mechanisms behind a common biofilm phenotype.
The evidence is clear: the combination of AFM and CLSM is not merely beneficial but imperative for advanced biofilm research. While AFM provides unparalleled insight into the nanomechanical and topographical properties that govern biofilm stability and adhesion, CLSM delivers an essential understanding of the three-dimensional spatial architecture and composition in a hydrated, near-native state. Their correlative application directly addresses the inherent limitations of each standalone technique, enabling researchers to build robust, quantitative structure-property relationships. As demonstrated by the latest research in large-area AFM and machine learning, the continued evolution of this correlative paradigm will be pivotal in unraveling the complex dynamics of biofilms and developing effective strategies to control them in medical, industrial, and environmental settings.
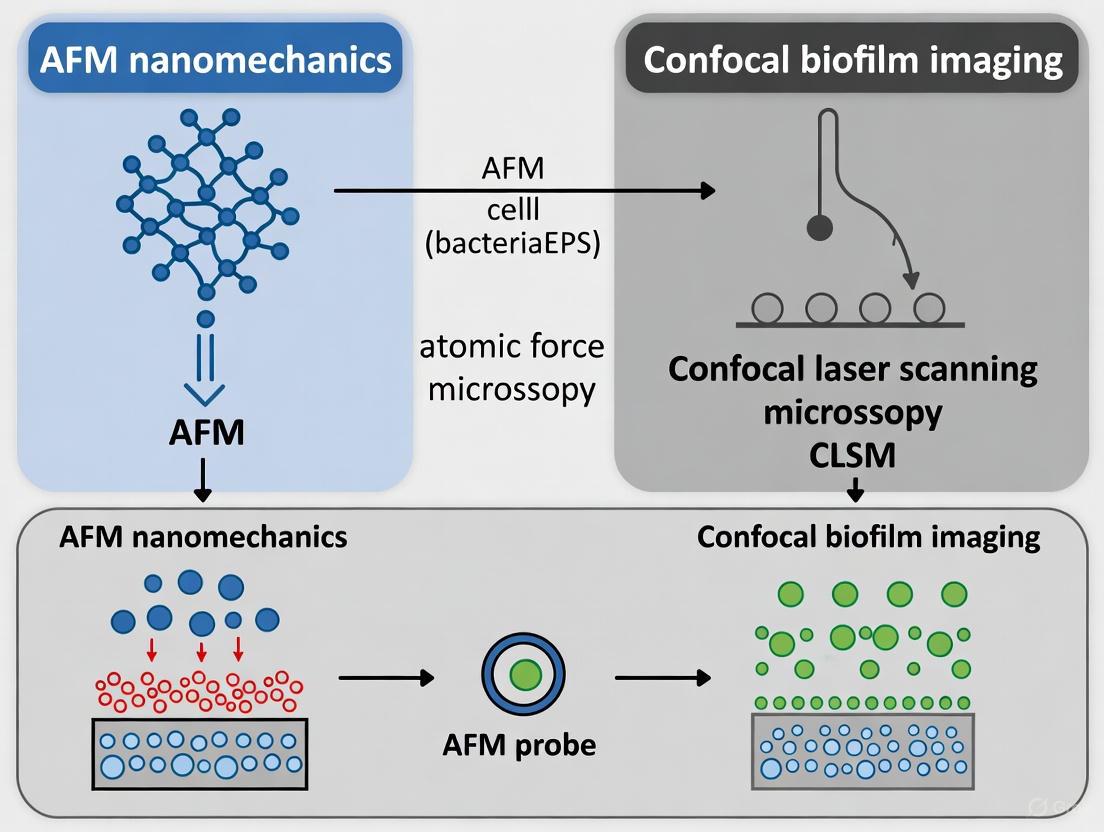
A Practical Workflow for Correlative AFM-Confocal Microscopy in Biofilm Analysis
Sample Preparation Strategies for Multimodal Imaging
The integration of Atomic Force Microscopy (AFM) with confocal laser scanning microscopy (CLSM) represents a powerful multimodal platform for biofilm research, enabling the correlation of nanoscale structural and mechanical properties with biochemical composition and viability data in a biological context [31] [2]. The primary challenge in such correlative studies lies in designing sample preparation protocols that simultaneously satisfy the distinct and often conflicting requirements of each technique without compromising the biofilm's native state. AFM demands relatively clean, fixed, or dry samples for optimal nanomechanical mapping and high-resolution topographical imaging, typically requiring substrates that are flat, rigid, and conductive [7] [32]. In contrast, CLSM relies on optical transparency and frequently involves hydrated, fluorescently stained samples to visualize internal structures and biological activity in three dimensions [2] [33]. This article objectively compares sample preparation strategies for standalone and correlated imaging, providing a structured guide to help researchers navigate the methodological trade-offs. By presenting standardized protocols and quantitative data on their outcomes, we aim to facilitate robust experimental design for correlating AFM nanomechanics with confocal biofilm imaging.
Comparative Analysis of Imaging Techniques and Their Sample Requirements
A fundamental understanding of each technique's operating principles and limitations is a prerequisite for designing effective multimodal sample preparation. The following table summarizes the core requirements for AFM, CLSM, and their integrated application.
Table 1: Core Requirements for AFM, CLSM, and Multimodal Imaging
| Imaging Technique | Spatial Resolution | Key Outputs | Ideal Sample State | Critical Substrate Properties |
|---|---|---|---|---|
| Atomic Force Microscopy (AFM) | ~0.1 nm laterally, ~0.01 nm vertically [31] | 3D topography, nanomechanical properties (stiffness, adhesion, viscoelasticity) [32] | Clean, fixed, or dry; can be in liquid [7] | Flat, rigid, low roughness (e.g., glass, mica, silicon wafers) [7] |
| Confocal Laser Scanning Microscopy (CLSM) | ~200 nm laterally, ~500 nm axially [33] | 3D morphology, biochemical localization, cell viability (via live/dead staining) [2] | Hydrated, fluorescently stained (live or fixed) [33] | Optically transparent (e.g., glass coverslips) [33] |
| Correlative AFM-CLSM | Combines nanoscale (AFM) and sub-diffraction (CLSM) resolution | Links nanomechanics with biochemical identity and spatial distribution [31] [2] | Compromise: Fixed and stained, but may be hydrated or dry for sequential imaging | Must satisfy both techniques: optically transparent, flat, and rigid [31] |
Sample Preparation Workflows: A Comparative Guide
Sample preparation strategies can be broadly categorized by the final state of the biofilm, which directly dictates the type of information that can be obtained. The choice of protocol involves critical trade-offs between preserving native structure, maintaining biological activity, and achieving technical compatibility.
Table 2: Comparison of Sample Preparation Protocols for Biofilm Imaging
| Preparation Strategy | Key Steps (Simplified) | Compatibility | Impact on Data | Best For |
|---|---|---|---|---|
| Minimal Preparation (Native State) | 1. Grow biofilm on suitable substrate (e.g., glass).2. Gently rinse with buffer to remove planktonic cells.3. Image in liquid [7] [31]. | AFM (in liquid): HighCLSM (live): HighCorrelative: High | Preserves native structure & mechanics; allows dynamic studies. | Investigating live biofilm processes, real-time antibiotic effects [31]. |
| Chemical Fixation | 1. Grow and rinse biofilm.2. Fix with aldehyde (e.g., 2.5% glutaraldehyde, 4% PFA).3. Rinse with buffer [2]. | AFM: HighCLSM (fixed): HighCorrelative: High | Stabilizes structure; may alter nanomechanical properties. | Preserving spatial relationships for high-resolution structural correlation. |
| Chemical Fixation & Staining | 1. Fix biofilm.2. Permeabilize (if needed).3. Apply fluorescent stains (e.g., FISH, FITC-ConA).4. Rinse and mount [2] [33]. | AFM: Medium*CLSM: EssentialCorrelative: Essential | Enables biochemical specificity in CLSM; risk of sample contamination or topography alteration for AFM. | Linking nanomechanics with specific molecular components (e.g., EPS). |
| Dehydration & Drying | 1. Fix biofilm.2. Dehydrate with graded ethanol or acetone series.3. Critical point drying or air-drying [7]. | AFM: HighCLSM: Low/NoneCorrelative: Challenging | Causes significant structural collapse; not suitable for most CLSM. | High-resolution AFM topography where hydration is not a concern [7]. |
*AFM on stained samples is possible but requires caution to avoid contaminating the tip with fluorescent dyes.
Detailed Experimental Protocol for Correlative AFM-CLSM
The following protocol is adapted from methodologies used in recent studies for investigating early bacterial attachment and biofilm formation, suitable for correlating nanomechanics with confocal data [7] [2].
Methodology: Sample Preparation for Fixed Correlative AFM-CLSM Imaging
Key Research Reagent Solutions:
- Substrate: PFOTS-treated glass coverslips or bare glass coverslips (#1.5 thickness, 25-35 mm diameter). The PFOTS treatment creates a hydrophobic surface that can modulate bacterial adhesion [7].
- Fixative Solution: Phosphate Buffered Saline (PBS) containing 2.5% glutaraldehyde.
- Staining Solution: PBS containing fluorescent stains (e.g., SYTO 9 for nucleic acids, FITC-labeled Concanavalin A for polysaccharides, propidium iodide for dead cells), with concentrations as per manufacturer's guidelines.
- Wash Buffer: Sterile physiological buffer (e.g., 10 mM PBS or 10 mM HEPES, pH 7.4).
Procedure:
- Substrate Preparation: Place a sterile glass coverslip in a petri dish or flow cell. For hydrophobic surfaces, treat with PFOTS or similar silane [7].
- Biofilm Growth: Inoculate with the bacterial strain of interest (e.g., Pantoea sp. YR343) in an appropriate liquid growth medium. Incubate for the desired duration to form early-stage biofilms or microcolonies [7].
- Gentle Rinsing: Carefully remove the substrate from the growth medium and immerse it in wash buffer to remove non-adherent planktonic cells. Avoid high shear forces that could disrupt delicate surface attachments.
- Chemical Fixation: Immerse the sample in the glutaraldehyde fixative solution for a minimum of 1 hour at 4°C. This step cross-links and stabilizes the biofilm structure.
- Buffer Rinse: Rinse the fixed sample three times (5 minutes each) with wash buffer to remove excess fixative.
- Fluorescent Staining: Incubate the sample with the chosen staining solution in the dark for 15-30 minutes.
- Final Rinse: Rinse the sample gently with wash buffer to remove unbound stain.
- Mounting for Imaging: For sequential imaging, the sample can be air-dried for AFM first, then mounted for CLSM. For simultaneous imaging, keep the sample hydrated and mount in a liquid cell.
Experimental Workflow and Data Correlation Logic
The following diagram illustrates the logical pathway and key decision points for conducting a correlative AFM-CLSM study on biofilms, from sample preparation through data analysis.
The strategic preparation of biofilm samples is the most critical determinant of success in multimodal studies seeking to correlate AFM-derived nanomechanics with confocal microscopy imaging. No single protocol is universally superior; the choice must be guided by the specific research question, whether it demands the preservation of live dynamics or the stabilized, stained structure for compositional analysis. As the field advances, the integration of machine learning for image stitching and analysis [7], coupled with more sophisticated correlative workflows, will further minimize the compromises inherent in sample preparation. By objectively evaluating the trade-offs outlined in this guide, researchers can make informed decisions that enhance the reliability and biological relevance of their multimodal data, ultimately driving a deeper understanding of biofilm architecture and function.
The study of biofilms, complex microbial communities encased in extracellular polymeric substances (EPS), requires analytical methods that can capture both their three-dimensional architecture and their mechanical properties [7] [1]. While Confocal Laser Scanning Microscopy (CLSM) excels at visualizing the spatial organization and biochemical characteristics of biofilms through optical sectioning and fluorescence, Atomic Force Microscopy (AFM) provides complementary nanoscale topographical imaging and quantitative mechanical property mapping [34] [35]. Individually, each technique offers valuable but incomplete insights; together, they enable researchers to correlate structural features with mechanical function. This guide examines the sequential integration of 3D CLSM mapping with AFM nanomechanics, providing researchers with a comprehensive framework for studying biofilm organization, resilience, and response to therapeutic interventions.
The limitations of single-technique approaches become particularly evident when studying heterogeneous structures like biofilms. CLSM provides exceptional volumetric imaging of hydrated, living biofilms but lacks the resolution to visualize individual bacterial appendages or measure mechanical properties [1]. Conversely, AFM achieves nanometer-scale resolution and can quantify elastic modulus, adhesion, and viscoelastic properties, but typically examines small surface areas that may not represent broader biofilm architecture [7] [35]. The correlative approach bridges this scale gap, enabling researchers to identify regions of interest with CLSM and subsequently characterize their nanomechanical properties with AFM. This protocol is particularly valuable for investigating structure-function relationships in biofilms, which exhibit remarkable spatial heterogeneity in both composition and mechanical behavior [7] [1].
Technical Comparison of Imaging Modalities
Fundamental Principles and Capabilities
Confocal Laser Scanning Microscopy (CLSM) operates on the principle of optical sectioning, where a pinhole eliminates out-of-focus light, allowing high-resolution imaging at specific depths within a sample [34]. By collecting sequential z-plane images, CLSM reconstructs three-dimensional representations of biofilms, typically with a lateral resolution of approximately 200 nm and axial resolution of 500-700 nm. CLSM predominantly provides qualitative and spatial information about biofilm architecture, matrix distribution, and cellular organization through fluorescent labeling techniques [1] [34].
Atomic Force Microscopy (AFM) functions by scanning a sharp probe attached to a flexible cantilever across a sample surface while monitoring cantilever deflection [36]. AFM captures topographical images with atomic-scale resolution and simultaneously quantifies mechanical properties through force-distance curves [35] [36]. Unlike CLSM, AFM does not require fluorescent labeling or conductive coatings, enabling analysis under near-physiological conditions [36]. Advanced AFM modes like force volume, nano-DMA, and parametric methods generate spatially-resolved maps of mechanical properties including elastic modulus, adhesion, and viscoelasticity [35].
Table 1: Fundamental Characteristics of CLSM and AFM
| Parameter | Confocal Laser Scanning Microscopy (CLSM) | Atomic Force Microscopy (AFM) |
|---|---|---|
| Resolution | Lateral: ~200 nm, Axial: ~500-700 nm | Sub-nanometer (vertical), Nanometer (lateral) |
| Imaging Depth | Tens to hundreds of micrometers | Surface topology (nanometers) |
| Sample Environment | Physiological conditions possible | Physiological liquids, air, vacuum |
| Key Measurements | 3D architecture, fluorescence localization, viability | Topography, elastic modulus, adhesion, surface roughness |
| Sample Preparation | Fluorescent staining, minimal processing | Often minimal, no labeling required |
| Throughput | Relatively fast (minutes for 3D stack) | Slow (typically minutes to hours per image) |
| Live Cell Imaging | Excellent with appropriate environmental control | Possible but challenging due to scanning speed |
Quantitative Performance Comparison
The complementary nature of CLSM and AFM becomes evident when examining their quantitative outputs. CLSM excels at measuring volumetric parameters such as biofilm thickness, biovolume, surface area-to-volume ratios, and spatial distribution of specific fluorescent markers [1]. AFM provides nanomechanical data including Young's modulus (typically 0.1-100 kPa for biological samples), adhesion forces (pN to nN range), and viscoelastic parameters [35] [36]. When applied sequentially, these techniques can establish correlations between biofilm localization patterns and their mechanical properties, offering insights into how structural organization influences function.
Table 2: Measurement Capabilities and Experimental Data Outputs
| Measurement Type | CLSM Capabilities | AFM Capabilities | Representative Experimental Data |
|---|---|---|---|
| Spatial Resolution | ~200 nm lateral, ~500 nm axial | <1 nm vertical, <5 nm lateral | AFM resolves individual flagella (~20-50 nm height) [7] |
| Structural Imaging | 3D architecture, matrix distribution, microcolonies | Surface topography, cellular morphology, appendages | AFM visualizes honeycomb patterns in Pantoea sp. biofilms [7] |
| Mechanical Properties | Limited to indirect inferences | Direct quantification of elastic modulus, adhesion | Young's modulus maps with nanoscale resolution [35] [36] |
| Chemical Specificity | Excellent with fluorescent labeling | Limited without functionalized tips | CLSM localizes specific EPS components [1] |
| Environmental Flexibility | Physiological conditions, live cell imaging | Air, liquid, controlled atmospheres | AFM measures live cells under physiological conditions [36] |
| Throughput | Moderate to high (3D stacks in minutes) | Low (typically slow scanning) | Large-area automated AFM addresses throughput limitations [7] |
Sequential Imaging Protocol Workflow
Experimental Design and Sample Preparation
The successful integration of CLSM and AFM requires careful experimental planning to maintain sample compatibility between instruments. Biofilms should be grown on substrates suitable for both techniques, typically glass-bottom dishes or coverslips that accommodate high-resolution oil immersion objectives for CLSM and provide sufficiently smooth surfaces for AFM topography scanning [34]. For live cell imaging, environmental control must be maintained throughout the process to preserve biofilm viability and native structure.
The sample preparation protocol begins with culturing biofilms under relevant conditions. For the Conpokal method (combined confocal and AFM), researchers recommend turning on instrument power sources at least one hour before experiments to ensure thermal equilibration [34]. Biofilms are typically fixed in a solution such as phosphate-buffered saline (PBS) or another clear liquid with low autofluorescence to prevent interference with fluorescence detection [34]. For bacterial samples like Streptococcus mutans, preparation involves inoculation in appropriate media overnight, followed by coating experimental dishes with poly-L-lysine solution to enhance surface adhesion approximately one hour before incubation completion [34].
Diagram 1: Sequential Imaging Workflow from CLSM to AFM. This workflow ensures correlative data collection from the same biofilm regions.
CLSM Imaging Protocol
The CLSM imaging phase begins with locating regions of interest within the biofilm sample. For structural assessment, collect z-stacks with appropriate step sizes (typically 0.5-1 μm) to resolve three-dimensional architecture. Optimize laser power, gain, and pinhole size to maximize signal-to-noise ratio while minimizing photobleaching and phototoxicity [34]. When imaging multiple fluorescence channels, sequential scanning prevents bleed-through between channels. For time-lapse experiments investigating biofilm dynamics, ensure environmental control throughout imaging.
CLSM settings must balance resolution requirements with sample viability. Higher numerical aperture objectives (60x or 100x oil immersion) provide superior resolution for detailed structural analysis but reduce working distance, potentially limiting thick biofilm imaging [1] [34]. The resolution advantage of CLSM over conventional light microscopy is particularly evident when imaging complex biofilm architectures, where its optical sectioning capability enables precise three-dimensional reconstruction of microbial communities and extracellular matrix components [1].
Region of Interest Transfer and AFM Integration
Following CLSM characterization, the transfer of specific regions of interest to the AFM represents a critical step in the correlative workflow. Maintain careful coordinate tracking of imaged areas, particularly when using motorized stages with positional encoding. For the Conpokal method that integrates both instruments, this transfer occurs within the same platform, eliminating registration challenges [34]. With separate instruments, fiduciary markers on substrates facilitate relocating the same areas.
Before AFM scanning, select appropriate cantilevers based on experimental requirements. Softer cantilevers (spring constants 0.01-0.1 N/m) are suitable for biological samples to minimize deformation, while stiffer cantilevers (0.5-1 N/m) provide better stability for topographic imaging [34] [35]. Calibrate each cantilever's spring constant and sensitivity using thermal tune or force curve methods on a rigid reference surface (e.g., clean glass or silicon) in the same medium used for biofilm imaging [34].
AFM Nanomechanical Mapping
AFM nanomechanical characterization employs several operational modes, each with distinct advantages for biofilm analysis. Force volume mode acquires force-distance curves at each pixel, providing the most comprehensive mechanical dataset but requiring extended acquisition times [35]. This approach enables quantitative mapping of elastic modulus through contact mechanics models like Hertz, Sneddon, or JKR theories [35] [36]. For heterogeneous biofilms, force volume mapping reveals mechanical variations correlated with structural features identified by CLSM.
PeakForce Tapping and other quantitative nanomechanical mapping (QNM) modes offer faster mechanical property characterization by performing force curves at kilohertz frequencies, significantly reducing acquisition time while maintaining nanoscale resolution [35]. These methods are particularly valuable for capturing dynamic processes in biofilms or surveying larger areas to contextualize CLSM observations. Additionally, nano-DMA (Dynamic Mechanical Analysis) modes characterize viscoelastic properties through oscillatory testing, revealing time-dependent mechanical behavior relevant to biofilm response under fluid flow or mechanical stress [35].
Research Reagent Solutions and Essential Materials
Table 3: Essential Research Reagents and Materials for Correlative CLSM-AFM
| Item | Function/Application | Examples/Specifications |
|---|---|---|
| Glass-bottom Dishes | Substrate for imaging | #1.5 thickness (0.16-0.19 mm) for high-resolution objectives |
| Poly-L-lysine | Surface coating for enhanced cell adhesion | 0.1% w/v aqueous solution, incubate 1 hour before use [34] |
| Fluorescent Stains | CLSM visualization of biofilm components | SYTO dyes for nucleic acids, ConA for polysaccharides, FITC for proteins |
| Phosphate Buffered Saline (PMS) | Imaging buffer | Maintains physiological pH and osmolarity, low autofluorescence [34] |
| AFM Cantilevers | Nanomechanical probing | Silicon nitride tips, spring constants 0.01-1 N/m (e.g., MLCT, PNP-TR) |
| Fixation Reagents | Sample preservation (optional) | Glutaraldehyde, formaldehyde (may affect mechanical properties) |
| Calibration Samples | AFM cantilever verification | Polystyrene beads, clean silicon wafer, gratings |
Applications in Biofilm Research and Therapeutic Development
The sequential CLSM-to-AFM protocol enables sophisticated analysis of biofilm responses to antimicrobial agents, surface modifications, and other interventions. CLSM can track changes in biofilm viability, thickness, and matrix composition following treatment, while subsequent AFM characterization quantifies associated mechanical alterations [1]. This approach is particularly valuable for evaluating anti-biofilm strategies including CRISPR-Cas-modified bacteriophages, quorum-sensing inhibitors, and enzyme-functionalized nanocarriers [1].
Recent advances in large-area automated AFM address historical limitations in scan range, enabling correlation between CLSM-identified features and nanomechanical properties across millimeter-scale areas [7]. When integrated with machine learning for image stitching and analysis, this approach reveals previously obscured heterogeneities in biofilm organization, such as the distinctive honeycomb pattern and preferred cellular orientation observed in Pantoea sp. YR343 biofilms [7]. These structural patterns directly influence mechanical properties and functional characteristics including antibiotic penetration resistance.
Diagram 2: Correlative Data Integration and Research Applications. Combined CLSM-AFM data enables both fundamental research and therapeutic development.
The sequential integration of 3D CLSM mapping with AFM nanomechanics represents a powerful methodological approach for investigating complex biofilm systems. This correlative protocol enables researchers to bridge critical scale gaps between micron-level architectural organization and nanometer-scale mechanical properties, providing unprecedented insights into structure-function relationships microbial communities. As both imaging technologies continue to advance—with developments in large-area AFM, machine learning-enhanced analysis, and more sophisticated correlative platforms—this sequential imaging approach will play an increasingly important role in both fundamental biofilm research and therapeutic development aimed at combating biofilm-associated infections.
Atomic Force Microscopy (AFM) has established itself as a pivotal tool in life sciences for quantifying the nanomechanical properties of biological systems, including bacterial biofilms. This guide provides a systematic comparison of AFM methodologies for measuring three fundamental parameters—stiffness, adhesion, and cohesive energy—within the context of biofilm research. The ability to correlate these AFM-derived nanomechanical properties with data from confocal laser scanning microscopy (CLSM) offers a powerful multi-modal approach for understanding biofilm structure-function relationships. Where CLSM reveals architectural organization and chemical composition of the extracellular polymeric substance (EPS), AFM quantifies the resulting mechanical properties that influence biofilm stability, virulence, and resistance to treatment [1]. This integrated framework enables researchers to connect microscale structural heterogeneity with nanoscale mechanical behavior, providing critical insights for pharmaceutical development targeting biofilm-associated infections.
Quantitative Comparison of AFM-Measured Parameters
Stiffness and Elastic Modulus Measurements
The stiffness of biofilm components is typically expressed through Young's modulus (E), a fundamental mechanical property calculated from AFM force-indentation curves using contact mechanics models, most commonly Sneddon's variations of the Hertz model [37].
Table 1: Representative Stiffness Values from AFM Biofilm Studies
| Material/Biofilm Component | Young's Modulus (E) | Measurement Conditions | AFM Technique | Citation |
|---|---|---|---|---|
| P. aeruginosa PAO1 (early biofilm) | Instantaneous Elastic Modulus: ~0.5 GPa (estimated from data) | Native conditions, microbead force spectroscopy | Voigt Standard Linear Solid model | [38] |
| P. aeruginosa PAO1 (mature biofilm) | Instantaneous Elastic Modulus: Reduced vs. early biofilm | Native conditions, microbead force spectroscopy | Voigt Standard Linear Solid model | [38] |
| P. aeruginosa wapR mutant (early biofilm) | Instantaneous Elastic Modulus: Drastically reduced vs. PAO1 | Native conditions, microbead force spectroscopy | Voigt Standard Linear Solid model | [38] |
| Polymer Reference (LDPE) | 0.20 GPa → 0.11 GPa (25°C → 100°C) | Temperature-controlled stage, PinPoint mode | Sneddon-Hertz model (conical indenter) | [39] |
| Polymer Reference (PVDF) | 2.50 GPa → 1.10 GPa (25°C → 100°C) | Temperature-controlled stage, PinPoint mode | Sneddon-Hertz model (conical indenter) | [39] |
Adhesion Force Measurements
Adhesion forces quantified by AFM reflect the sum of attractive interactions between the AFM probe and the sample surface, including van der Waals forces, electrostatic interactions, and polymer bridging [40].
Table 2: Representative Adhesion Force Values from AFM Biofilm Studies
| Interaction Pair | Adhesion Force | Adhesive Pressure | Measurement Conditions | AFM Technique |
|---|---|---|---|---|
| E. coli TG1 Goethite | -97 ± 34 pN (initial attractive force) | Not reported | Water, cell probe on mineral surface | Single-cell force spectroscopy |
| E. coli TG1 Goethite (after 4s) | -3.0 ± 0.4 nN (maximum adhesion) | Not reported | Water, cell probe on mineral surface | Single-cell force spectroscopy |
| P. aeruginosa PAO1 (early biofilm) Glass | Not reported | 34 ± 15 Pa | Standardized conditions, defined contact area | Microbead Force Spectroscopy (MBFS) |
| P. aeruginosa wapR (early biofilm) Glass | Not reported | 332 ± 47 Pa | Standardized conditions, defined contact area | Microbead Force Spectroscopy (MBFS) |
| P. aeruginosa PAO1 (mature biofilm) Glass | Not reported | 19 ± 7 Pa | Standardized conditions, defined contact area | Microbead Force Spectroscopy (MBFS) |
| P. aeruginosa wapR (mature biofilm) Glass | Not reported | 80 ± 22 Pa | Standardized conditions, defined contact area | Microbead Force Spectroscopy (MBFS) |
| Si₃N₄ Tip SRB Cell Body | -3.9 to -4.3 nN | Not reported | Seawater on mica | Force-distance curve |
| SRB Cell Substratum Periphery | -5.1 to -5.9 nN | Not reported | Seawater on mica | Force-distance curve |
| SRB Cell Cell Interface | -6.5 to -6.8 nN | Not reported | Seawater on mica | Force-distance curve |
Cohesive Energy Measurements
Cohesive energy represents the energy required to separate biofilm constituents and is a direct indicator of the internal strength of the EPS matrix.
Table 3: Cohesive Energy Measurements from AFM Biofilm Studies
| Biofilm Type | Cohesive Energy | Measurement Conditions | AFM Technique |
|---|---|---|---|
| Activated Sludge (1-day biofilm) | 0.10 ± 0.07 to 2.05 ± 0.62 nJ/μm³ | 90% humidity, moist biofilm | Scan-induced abrasion |
| Activated Sludge (+10 mM Ca²⁺) | 0.10 ± 0.07 to 1.98 ± 0.34 nJ/μm³ | 90% humidity, moist biofilm | Scan-induced abrasion |
Experimental Protocols for Key AFM Measurements
Microbead Force Spectroscopy (MBFS) for Adhesion and Viscoelasticity
This protocol, adapted from Abu-Lail & Camesano (2009), enables simultaneous quantification of adhesive and viscoelastic properties under standardized conditions [38].
- Probe Functionalization: Attach a ~50 µm diameter glass bead to a tipless AFM cantilever using a suitable epoxy. Sterilize the probe if needed.
- Biofilm Coating: Incubate the bead-coated cantilever with a concentrated suspension of the bacterial strain (e.g., OD₆₀₀ = 2.0 for P. aeruginosa) to form a monolayer of cells on the bead.
- System Calibration: Calibrate the cantilever's spring constant using the thermal tune method. Precisely determine the photodetector sensitivity.
- Standardized Force Measurement: Engage the biofilm-coated probe with a clean, sterile glass substrate in liquid. Use standardized contact parameters: set point force, contact time (e.g., 0.5-2 seconds), and retraction speed.
- Data Collection: Record at least 100-200 force-distance curves at multiple locations on the substrate to ensure statistical significance.
- Adhesion Analysis: Calculate the adhesive pressure from the retraction force curves by dividing the maximum adhesion force by the contact area.
- Viscoelastic Analysis: Fit the indentation-versus-time data (from the "hold" segment at constant force) to a viscoelastic model (e.g., Voigt Standard Linear Solid model) to extract elastic moduli and viscosity.
AFM Abrasion Test for Cohesive Energy
This protocol, based on Ahimou et al. (2007), directly measures the cohesive strength within a biofilm by quantifying the energy required to remove material via AFM tip abrasion [41].
- Biofilm Preparation: Grow a young biofilm (e.g., 1-day old) on a flat, rigid substrate (e.g., a gas-permeable membrane).
- Humidity Control: Equilibrate the hydrated biofilm in the AFM chamber at a constant, high relative humidity (e.g., 90%) to maintain consistent water content.
- Baseline Imaging: Acquire a high-resolution topographic image of a selected region (e.g., 5x5 µm) at a minimal applied load (~0 nN) to avoid sample damage.
- Controlled Abrasion: Zoom into a smaller sub-region (e.g., 2.5x2.5 µm) and perform repeated raster scans with an elevated load (e.g., 40 nN) to abrade the biofilm.
- Post-Abrasion Imaging: Return to the low load and re-image the original 5x5 µm area to capture the abraded crater.
- Volume and Energy Calculation:
- Calculate the volume of removed biofilm by subtracting the post-abrasion topography from the pre-abrasion topography.
- Determine the total frictional energy dissipated during abrasion from the lateral (friction) signal of the AFM photodiode.
- Calculate the cohesive energy (Γ) using the formula: Γ = (Total Frictional Energy Dissipated) / (Volume of Biofilm Removed). The unit is J/m³ or, equivalently, nJ/µm³.
Force Volume Mapping for Spatial Stiffness Variations
This protocol is used to create quantitative maps of Young's modulus, revealing mechanical heterogeneity across a biofilm [35].
- Probe Selection: Choose a cantilever with a well-defined tip geometry (e.g., spherical or conical) and a known spring constant.
- Grid Definition: Define a grid of measurement points (e.g., 64x64 or 128x128) over the area of interest on the biofilm surface.
- Force-Distance Curve Acquisition: At each point in the grid, perform a complete force-distance cycle (approach and retract). This collection of curves is the "force volume" data set.
- Data Processing: For each approach curve, identify the point of contact and the region of indentation.
- Model Fitting: Fit the indentation segment of each curve to an appropriate contact mechanics model (e.g., Sneddon-Hertz model for a conical tip). The model relates the applied force (F) to the indentation depth (δ) and Young's modulus (E).
- Map Generation: Create a spatial map where the color at each pixel corresponds to the calculated Young's modulus value from the force curve at that location.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 4: Key Reagents and Materials for AFM Biofilm Nanomechanics
| Item | Function/Application | Example Specifications / Notes |
|---|---|---|
| AFM Cantilevers | Transducer for force measurement; different shapes for different measurements. | Tipless Cantilevers: For attaching microbeads or cells (MBFS). Sharp Tips (PSPN-type): For high-resolution imaging and force spectroscopy on single cells. Colloidal Probes: Spherical tips for defined contact geometry. |
| Functionalization Reagents | To glue particles or immobilize cells/biofilms on cantilevers and substrates. | Epoxy: For attaching microbeads. Poly-L-Lysine: As an adhesive coating to immobilize cells on substrates. Glutaraldehyde: As a crosslinker for stronger cell fixation (alters native properties). |
| Standardized Substrates | Provide a consistent, well-defined surface for adhesion and cohesion measurements. | Glass Microspheres/Coverslips: For adhesion pressure measurements in MBFS. Gas-Permeable Membranes: For growing biofilms under controlled aeration. Mica: Provides an atomically flat surface for single-cell studies. |
| Calibration Materials | Essential for verifying the spring constant of cantilevers and the sensitivity of the photodetector. | Clean, rigid sample (e.g., silicon wafer): For sensitivity calibration. Cantilever thermal tune method: Standard for in-situ spring constant calibration. |
| Viscoelastic Model | Mathematical framework to extract quantitative mechanical properties from raw data. | Voigt Standard Linear Solid Model: A common model used to fit creep data and extract instantaneous/delayed elastic moduli and viscosity [38]. |
| Contact Mechanics Model | Mathematical framework to calculate Young's Modulus from force-indentation data. | Sneddon-Hertz Model: The most widely used model for analyzing indentation of an elastic sample by a conical or spherical indenter [37]. |
Workflow for Correlative AFM and Confocal Biofilm Imaging
The integration of AFM nanomechanics with confocal imaging creates a powerful correlative workflow. The following diagram illustrates the sequential steps to acquire complementary structural and mechanical data from the same biofilm sample.
Correlative AFM-Confocal Biofilm Analysis Workflow
Integrated Framework for Data Correlation
The ultimate goal of a multi-modal approach is to establish quantitative relationships between the structural features visualized by confocal microscopy and the mechanical properties measured by AFM. The following diagram outlines the logical framework for integrating these datasets.
Framework for CLSM-AFM Data Correlation
Confocal Laser Scanning Microscopy (CLSM) has emerged as a cornerstone technique in biofilm research, enabling non-destructive, three-dimensional visualization and quantification of complex biofilm architectures. Unlike traditional methods such as crystal violet staining or colony-forming unit counts, which provide limited structural information, CLSM allows researchers to precisely extract critical parameters including biofilm thickness, biovolume, and surface coverage under physiological conditions [1] [42]. This capability is particularly valuable when correlating structural data with nanomechanical properties obtained from Atomic Force Microscopy (AFM), creating a comprehensive picture of biofilm behavior and response to interventions [28]. The transition from qualitative observation to quantitative analysis through CLSM represents a significant advancement in understanding biofilm organization, heterogeneity, and function across medical, industrial, and environmental contexts.
The inherent heterogeneity of biofilms—characterized by variations in cellular density, extracellular polymeric substance (EPS) distribution, and void spaces—poses a significant challenge for accurate analysis [43]. CLSM addresses this challenge by providing high-resolution z-stack images that capture the full three-dimensional complexity of biofilms, enabling researchers to move beyond two-dimensional approximations and derive statistically robust measurements that reflect true biofilm organization [44] [42]. This article provides a comprehensive comparison of CLSM methodologies for extracting structural biofilm data, detailing experimental protocols, analytical approaches, and integration with complementary techniques like AFM.
Comparative Analysis of Biofilm Analysis Methods
Table 1: Comparison of Biofilm Analysis Techniques
| Method | Key Measurable Parameters | Resolution | Sample Preparation | Advantages | Limitations |
|---|---|---|---|---|---|
| CLSM | Thickness, biovolume, surface coverage, roughness, viability | ~200 nm laterally, ~500 nm axially [28] | Staining required, minimal disturbance | Non-destructive, 3D visualization, in situ analysis, viability assessment | Requires fluorescent markers; limited by stain penetration in thick biofilms |
| Atomic Force Microscopy (AFM) | Topography, mechanical properties (Young's modulus, adhesion) [28] | Sub-nanometer vertical, nanometer lateral [45] | Can be used in liquid without staining | Nanoscale resolution, quantitative mechanical data, no staining required | Small scan area, slow imaging speed, potential sample damage |
| Scanning Electron Microscopy (SEM) | Surface morphology, cellular arrangement | 1-10 nm | Dehydration, fixation, coating required | Ultra-high resolution surface details | Destructive sample preparation, no live imaging |
| Optical Coherence Tomography (OCT) | Biofilm thickness, macro-scale structure | 1-10 μm [28] | Minimal, can image through transparent surfaces | Deep penetration, rapid imaging, no staining | Lower resolution, limited chemical specificity |
| Crystal Violet Staining | Total biomass | N/A | Fixation, staining, destaining | Simple, cost-effective, high-throughput | No 3D information, no viability data, destructive |
CLSM Experimental Protocol for Biofilm Analysis
Sample Preparation and Staining
Proper sample preparation is fundamental to obtaining accurate CLSM data. Biofilms are typically grown on suitable substrates such as glass coverslips, hydroxyapatite discs (for oral biofilms), or membrane surfaces depending on the research context [46] [27]. For viability assessment, the LIVE/DEAD BacLight Bacterial Viability Kit is widely employed, containing SYTO 9 and propidium iodide (PI) stains. The staining protocol involves gently washing the biofilm to remove non-adherent cells, then applying the staining solution (typically 1-2 μL of each stain in 1 mL of distilled water) and incubating for 15 minutes in the dark [42] [46]. After incubation, samples are rinsed to remove excess stain and mounted for imaging while maintaining hydration to preserve native biofilm architecture.
Image Acquisition Parameters
Optimal CLSM image acquisition requires careful parameter selection. For a Leica SP8 system, SYTO 9 is typically excited at 488 nm with emission collected between 500-545 nm, while PI is excited at 561 nm with emission collected between 615-745 nm [42] [46]. A 20x-63x objective is commonly used, with numerical aperture ≥0.9 preferred for improved resolution. Z-stack images should be collected with step sizes of 0.5-1 μm to adequately capture the entire biofilm thickness, with the number of slices determined by preliminary scans to identify the biofilm boundaries. Laser power should be optimized to ensure sufficient signal while avoiding photobleaching or detector saturation [44].
Quantitative Analysis Workflow
Table 2: Key Structural Parameters Extractable from CLSM Data
| Parameter | Definition | Calculation Method | Biological Significance |
|---|---|---|---|
| Biofilm Thickness | Maximum vertical dimension of biofilm | Distance from substrate to biofilm-liquid interface [44] | Influences diffusion gradients, antimicrobial penetration |
| Average Thickness | Mean vertical dimension | Average of local thickness measurements across entire biofilm [27] | Indicator of overall biofilm development |
| Biovolume | Total volume occupied by biomass | Sum of volume elements occupied by stained biomass [42] | Proportional to total cellular and matrix content |
| Surface Coverage | Horizontal area occupied by biofilm | Percentage of substrate surface covered by biofilm [27] | Measure of colonization extent |
| Surface Roughness | Textural heterogeneity | Coefficient of variation in biofilm height [27] | Indicator of structural heterogeneity, related to mass transfer |
| Viability Ratio | Proportion of live cells | Biovolume of green fluorescence / total biovolume [42] | Metabolic status and treatment efficacy |
Figure 1: CLSM Biofilm Analysis Workflow. This diagram illustrates the sequential steps from sample preparation to data validation in quantitative CLSM analysis of biofilms.
Advanced Analysis Techniques and Algorithms
Automated Image Analysis Approaches
Traditional manual analysis of CLSM data is time-consuming and subject to observer bias. Automated analysis methods have been developed to improve reproducibility and efficiency. The Biofilm Viability Checker, an open-source tool developed for Fiji/ImageJ, incorporates image pre-processing and automated thresholding to quantify viability and biomass from live/dead stained biofilms [42]. This approach demonstrated significantly lower coefficient of variation (4.24-11.5%) compared to traditional CFU counting (17.0-78.1%), highlighting its improved precision for quantitative analysis [42].
For thickness analysis, R-based algorithms enable processing of large CLSM datasets with minimal manual intervention. One published method analyzes fluorescence intensity profiles across z-positions to determine biofilm thickness automatically, processing over 19,000 individual cells to generate detailed property maps across surface areas [44]. This high-throughput approach is essential for capturing biofilm heterogeneity and obtaining statistically significant results.
3D Reconstruction and Visualization
Advanced CLSM analysis extends beyond basic parameter extraction to comprehensive 3D reconstruction of biofilm architecture. Software packages such as IMARIS, COMSTAT, and BiofilmQ enable detailed visualization and quantification of complex biofilm features including pore networks, channel structures, and microcolony organization [42] [46]. These structural elements significantly impact mass transfer processes within biofilms, influencing nutrient availability, waste removal, and antimicrobial penetration [43]. The ability to correlate these architectural features with functional properties represents a significant advancement in biofilm research.
Correlation with AFM Nanomechanics
Multi-scale Analysis Approach
Integrating CLSM with AFM creates a powerful multi-scale analytical platform that correlates structural organization with mechanical properties. While CLSM provides mesoscale architectural information, AFM complements these data with nanoscale mechanical characterization, including Young's modulus, adhesion forces, and viscoelastic properties [28]. This combined approach has revealed significant structure-property relationships in oral biofilms, demonstrating that increasing sucrose concentration decreases Young's modulus while increasing cantilever adhesion—findings that would be impossible to obtain with either technique alone [28].
Large-Area AFM and Machine Learning Integration
Recent advancements in automated large-area AFM have addressed the traditional limitation of small imaging areas, enabling correlation with CLSM over biologically relevant scales [45] [7]. When combined with machine learning algorithms for image stitching, cell detection, and classification, this approach can cover millimeter-scale areas while maintaining nanometer-scale resolution [7]. The integration of artificial intelligence facilitates seamless analysis of the high-volume, information-rich data generated by these complementary techniques, enabling comprehensive structure-function analysis of biofilms across multiple spatial scales [43] [7].
Figure 2: Multi-scale Correlation of CLSM and AFM Data. This diagram illustrates how structural data from CLSM and mechanical properties from AFM can be integrated to generate comprehensive biological insights into biofilm behavior.
Essential Research Reagent Solutions
Table 3: Key Research Reagents for CLSM Biofilm Analysis
| Reagent/Equipment | Function | Application Notes |
|---|---|---|
| SYTO 9 Stain | Green fluorescent nucleic acid stain for membranes with intact integrity [42] | Used at 1-2 μL/mL concentration; 15-minute incubation |
| Propidium Iodide (PI) | Red fluorescent stain for compromised membranes [42] | Distinguishes viable vs. non-viable cells; can bind extracellular DNA |
| Glass Coverslips | Transparent substrate for biofilm growth | Compatible with high-resolution oil immersion objectives |
| Hydroxyapatite Discs | Mineralized surface for oral biofilm studies [28] | Mimics tooth enamel composition |
| PVC Coupons | Substrate for environmental biofilm studies [27] | Represents plumbing and industrial surfaces |
| Artificial Saliva Growth medium for oral biofilms [28] | Represents in vivo conditions for dental plaque research | |
| Brain Heart Infusion (BHI) | Nutrient-rich growth medium [28] | Promotes rapid biofilm formation |
| Phosphate Buffered Saline (PBS) | Washing and imaging buffer | Maintains osmotic balance during imaging |
CLSM has revolutionized quantitative biofilm analysis by enabling non-destructive, three-dimensional characterization of structural parameters essential for understanding biofilm development, function, and response to interventions. The extraction of accurate thickness, biovolume, and surface coverage data provides critical insights into biofilm architecture and heterogeneity. When correlated with AFM-derived nanomechanical properties, these structural parameters contribute to comprehensive structure-function relationships that enhance our understanding of biofilm behavior across medical, industrial, and environmental contexts. As automated analysis algorithms and multi-modal imaging approaches continue to advance, CLSM will remain an indispensable tool in the biofilm researcher's arsenal, driving innovations in biofilm management and control strategies.
In the study of complex biological systems like biofilms, a significant challenge persists: the disconnect between nanoscale mechanical properties and the functional macroscale organization of these structures. Biofilms are inherently heterogeneous microbial communities, characterized by spatial and temporal variations in structure, composition, and mechanical properties [7]. Traditional analytical methods often fail to capture the full scope of this complexity, unable to link local subcellular and cellular scale changes to the evolution of larger functional architectures [7]. Atomic force microscopy (AFM) has emerged as a powerful tool for nanomechanical characterization, providing detailed insights into structural and functional properties at the cellular and even sub-cellular level [7]. However, its limited scan range and labor-intensive nature have historically restricted its ability to connect these smaller scale features to the macroscale organization that determines biofilm function in medical, industrial, and environmental contexts [7]. This guide objectively compares current AFM-based technologies and methodologies that address this scale integration challenge, providing researchers with experimental data and protocols for correlating local nanomechanics with macroscale structural analysis.
Comparative Analysis of AFM Modalities for Multiscale Characterization
The integration of AFM with other analytical techniques has generated multiple technological pathways for correlative analysis. Each approach offers distinct advantages and limitations for specific research applications, particularly in the context of biofilm research and nanomechanical mapping.
Table 1: Comparison of AFM-Based Correlative Imaging Platforms
| Technology Platform | Spatial Resolution | Key Strengths | Primary Limitations | Representative Systems |
|---|---|---|---|---|
| Large-Area Automated AFM | Nanoscale (cellular) to millimeter-scale [7] | Automated scanning over mm² areas; ML-based stitching; minimal sample prep [7] | Limited to surface properties; requires drying for biofilm imaging [7] | Custom systems with machine learning integration [7] |
| AFM-Confocal Correlation | Nanoscale (AFM) to sub-micron (confocal) [47] | Simultaneous topological, mechanical, and chemical data; real-time under physiological conditions [47] | Complex system alignment; higher instrumentation costs [48] | Bruker Dimension Nexus; Park FX200 [48] |
| AFM-Nanomechanical Fingerprinting (NMF) | Nanoscale mechanical mapping [49] | Quantifies tissue stiffness; tracks fibrosis progression/therapy response [49] | Limited to excised tissues; requires biopsy [49] | Molecular Imaging-Agilent PicoPlus AFM [49] |
| Wavelet-Based Multifrequency AFM | Nanoscale material differentiation [50] | Enhances contrast via harmonics; distinguishes materials without labels [50] | Complex data interpretation; specialized expertise needed [50] | Wavelet-transform AFM with AFM-ICE algorithm [50] |
Table 2: Performance Metrics for AFM-Based Biofilm Characterization
| Characterization Method | Quantitative Outputs | Sample Throughput | Data Richness | Implementation Complexity |
|---|---|---|---|---|
| Large-Area AFM | Cell count, confluency, orientation, flagellar mapping [7] | Medium (automation enabled) [7] | High (structural heterogeneity over mm areas) [7] | High (requires ML integration) [7] |
| Nanomechanical Fingerprinting | Young's modulus, adhesion forces, viscoelastic parameters [49] | Low (point-by-point measurement) [49] | Medium (quantitative mechanics only) [49] | Medium (standard AFM operation) [49] |
| AFM-ICE Contrast Enhancement | Harmonic amplitudes, material contrast indices [50] | High (post-processing algorithm) [50] | Medium (enhanced material differentiation) [50] | Low (software implementation) [50] |
| Correlative AFM-Fluorescence | Topography, mechanics, and chemical colocalization [48] [47] | Low to Medium (dependent on modality) [48] | Very High (multimodal data integration) [48] | Very High (system alignment and operation) [48] |
Experimental Protocols for Integrated Nanomechanical-Structural Analysis
Large-Area AFM for Biofilm Assembly Mapping
Objective: To characterize spatial heterogeneity and cellular morphology during early biofilm formation over millimeter-scale areas, capturing features previously obscured by conventional AFM's limited scan range [7].
Sample Preparation:
- Pantoea sp. YR343 cultures are grown in liquid growth medium [7].
- PFOTS-treated glass coverslips placed in petri dish are inoculated with growing cells [7].
- At selected time points (e.g., 30 minutes, 6-8 hours), coverslips are removed and gently rinsed to remove unattached cells [7].
- Samples are dried before AFM imaging [7].
Instrumentation and Parameters:
- Commercial AFM system capable of automated large-area scanning [7].
- Integration with machine learning algorithms for image stitching, cell detection, and classification [7].
- Scan areas: Multiple adjacent regions totaling millimeter-scale coverage [7].
- Resolution: High enough to visualize cellular features (∼2µm length, ∼1µm diameter) and flagellar structures (∼20-50nm height) [7].
Data Analysis:
- ML-based segmentation for automated extraction of parameters: cell count, confluency, cell shape, and orientation [7].
- Quantitative analysis of spatial patterns, including preferred cellular orientation and honeycomb pattern formation [7].
- Flagellar mapping to assess coordination and potential role in biofilm assembly beyond initial attachment [7].
Nanomechanical Fingerprinting for Tissue Fibrosis Assessment
Objective: To identify unique nanomechanical fingerprints (NMFs) that characterize fibrosis stages and monitor treatment response in pulmonary fibrosis [49].
Sample Preparation:
- Human lung biopsies from PF patients or bleomycin-induced murine PF models [49].
- Specimens immediately placed in ice-cold PBS supplemented with protease inhibitor cocktail [49].
- Samples immobilized on 35mm plastic culture dish with fast-drying epoxy glue [49].
- AFM measurements performed within 1-72 hours post-tissue removal to prevent stiffness alterations [49].
Instrumentation and Parameters:
- AFM System: Molecular Imaging-Agilent PicoPlus AFM [49].
- Cantilevers: Appropriate for soft biological samples (specific spring constants not detailed) [49].
- Measurement Mode: Force spectroscopy mapping across tissue surface [49].
- Environment: PBS with protease inhibitors at room temperature [49].
Data Analysis:
- Extraction of nanomechanical parameters (Young's modulus, adhesion) from force curves [49].
- Correlation with histopathological staining, polarized and second harmonic generation (SHG) microscopy [49].
- Real-time PCR analysis of collagen I expression [49].
- In silico classification using Support Vector Machine (SVM) to validate NMFs as diagnostic biomarkers [49].
Integrated Workflow for Correlative AFM-Biofilm Analysis
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Research Reagents and Materials for AFM-Based Correlative Studies
| Reagent/Material | Function/Application | Specific Examples |
|---|---|---|
| PFOTS-Treated Glass | Creates hydrophobic surface for controlled bacterial attachment and biofilm formation [7] | Used for Pantoea sp. YR343 biofilm studies in large-area AFM [7] |
| Protease Inhibitor Cocktail | Preserves native mechanical properties by preventing tissue degradation during AFM measurement [49] | Complete Mini (Roche Diagnostics) in PBS for lung tissue biopsies [49] |
| Bleomycin Sulfate | Induces pulmonary fibrosis in murine models for studying disease progression and treatment monitoring [49] | Derived from Streptomyces verticillus; 3 U/mL concentration for intratracheal administration [49] |
| Pirfenidone | Anti-fibrotic drug for treatment validation studies using nanomechanical fingerprints [49] | Esbriet (Roche); 500 mg/kg daily oral dose in murine models [49] |
| Fast-Drying Epoxy Glue | Immobilizes tissue specimens for AFM nanomechanical measurements without altering properties [49] | Two-component epoxy for securing biopsies to culture dishes [49] |
| DSPE-mPEG Encapsulated NPs | Multimodal imaging probes for correlative AFM and NIR-II fluorescence imaging [51] | RENPs@DSPE-mPEG for bone, vascular, and lymphatic imaging [51] |
Data Integration Architectures for Multimodal Imaging and Omics
The complexity of data generated through correlative AFM and imaging approaches necessitates robust data management infrastructures. Effective integration of multimodal data enables researchers to derive novel insights into biological processes that would not be apparent from individual techniques alone [52].
FAIR Data Management Architecture for Correlative Imaging
Emerging Methodologies and Future Directions
Machine Learning-Enhanced AFM Image Analysis
Artificial intelligence and machine learning are transforming AFM data acquisition and analysis across four key areas: sample region selection, scanning process optimization, data analysis, and virtual AFM simulation [7]. ML applications in AFM image processing include:
- Automated Segmentation and Classification: ML algorithms enable automated detection of cellular features, significantly reducing analysis time for large-area AFM scans of biofilms [7].
- Image Contrast Enhancement: Unsupervised learning approaches like AFM-Image Contrast Enhancement (AFM-ICE) integrate image fusion and histogram equalization to improve visualization of complex multilayer structures [50].
- Harmonic Signal Analysis: Wavelet-based techniques capture nonlinear cantilever responses, extracting material properties from higher-order oscillations without additional hardware [50].
Correlative AFM Platforms and Integration Methodologies
The future of correlative AFM-nanomechanics integration lies in the development of more sophisticated multimodal platforms:
- Industry-Level Automation: Park Systems' FX200 provides fully automated large-sample capability, improving accessibility for biological researchers [48].
- Interferometry-Enhanced Sensing: Oxford Instruments' Cypher Vero uses Quadrature Phase Differential Interferometry (QPDI) for more accurate tip displacement sensing, particularly beneficial for techniques like Piezoresponse Force Microscopy (PFM) [48].
- Integrated AFM-Fluorescence Systems: These systems combine nanoscale topographical information with optical and spectral chemical data, enabling the linking of properties at the nanoscale that was not previously possible [48].
Overcoming Technical Hurdles: Optimizing Your Correlative AFM-Confocal Workflow
Atomic Force Microscopy (AFM) has become an indispensable tool for characterizing the structural and mechanical properties of biological samples, particularly within the field of biofilm research. The ability to correlate AFM nanomechanics with confocal microscopy data provides a powerful multi-modal approach to understanding the architecture and function of these complex microbial communities. However, a significant challenge persists: the potential for AFM probes to induce sample damage, especially when imaging delicate, hydrated biological materials. This guide objectively compares the performance of different gentle AFM scanning modes and provides supporting experimental data and protocols to help researchers make informed decisions that preserve sample integrity without compromising data quality.
Comparative Analysis of Gentle AFM Modes for Hydrated Imaging
The following table summarizes the key characteristics, optimal use cases, and relative gentleness of the primary AFM modes used for imaging sensitive samples in hydrated conditions.
Table 1: Comparison of Gentle AFM Scanning Modes for Hydrated Biological Samples
| AFM Mode | Principle of Operation | Best for Imaging | Relative Gentleness | Key Advantages for Live Samples | Notable Limitations |
|---|---|---|---|---|---|
| Intermittent Contact (Tapping) Mode [53] | Tip oscillates and briefly contacts the sample, minimizing lateral forces. | Live cells, soft polymers, hydrated biofilms [53]. | High | Minimal shear forces; reduced sample disruption [53]. | Can still transfer energy to soft samples; requires careful amplitude control. |
| Force Volume / Force Spectroscopy [35] [53] [11] | Collects force-distance curves at each pixel to map mechanical properties. | Nanomechanical property mapping (e.g., stiffness, adhesion) [35] [11]. | Medium (point-by-point) | Provides quantitative mechanical data; direct correlation with confocal data [35]. | Very slow imaging speed; potential for local indentation damage. |
| High-Speed AFM (HS-AFM) [15] | Very fast scanning to capture dynamic processes in near-real-time. | Conformational dynamics of single molecules, protein machinery [15]. | Context-Dependent | Captures dynamics; can use very small amplitudes [15]. | Limited scan size; specialized, expensive equipment. |
| Nanomechanical Imaging (Bimodal/Multimodal) [35] [11] | Excites multiple cantilever eigenmodes simultaneously to extract properties. | Stiffness and viscoelasticity mapping of heterogeneous materials [35] [11]. | High (when optimized) | High-speed, high-resolution mechanical mapping with minimal invasion [35]. | Complex setup and data interpretation; requires expert knowledge. |
Quantitative Performance Data in Biofilm & Biological Imaging
The selection of an appropriate AFM mode must be guided by empirical performance data. The following table consolidates key metrics and findings from recent studies on soft and hydrated samples.
Table 2: Experimental Data and Performance Metrics from Selected Studies
| Sample Type | AFM Mode Used | Key Experimental Parameters | Quantitative Outcome | Reported Effect on Sample Integrity |
|---|---|---|---|---|
| Pantoea sp. YR343 Biofilm [7] | Large Area Automated AFM (in air, on dried samples) | Automated stitching of high-resolution images; ML for analysis [7]. | Resolved cellular morphology (~2 µm length) and flagella (~20-50 nm height) [7]. | Drying required; not for live hydration. Highlights capability for fine surface detail. |
| Lamellar Bone (Hydrated) [54] | Not specified (topography & mechanical mapping) | Hydrated state maintained during imaging and modulus measurement [54]. | Young's modulus lower in hydrated vs. dehydrated state; tessellation pattern visible only when hydrated [54]. | Hydrated imaging preserved native microstructure; dehydration caused irreversible topographical changes. |
| Live Mammalian Cells [35] [11] | Force Volume (with sinusoidal modulation) | Off-resonance excitation to reduce higher harmonics [35] [11]. | Clear identification of viscoelastic hysteresis in force-distance curves [35] [11]. | Enabled property mapping of living cells without reported damage, under physiological conditions. |
| Single Proteins (e.g., TRPV3) [15] | High-Speed AFM (HS-AFM) | Near-physiological conditions, single-molecule resolution [15]. | Captured conformational dynamics at sub-second frame rates [15]. | Successfully imaged single molecules without apparent disruption of function. |
Experimental Protocols for Gentle AFM in Hydrated Biofilm Research
To achieve reproducible and damage-free imaging, a standardized experimental methodology is crucial. Below is a detailed protocol for nanomechanical characterization of hydrated biofilms, adaptable for correlation with confocal imaging.
Protocol 1: Hydrated Nanomechanical Mapping of Biofilms
This protocol is adapted from general guidelines for soft matter and specific applications in biological AFM [53] [54].
Sample Preparation:
- Grow biofilms on suitable substrates (e.g., glass coverslips, plastic dishes) compatible with both AFM and confocal microscopy.
- Gently rinse with appropriate buffer (e.g., PBS, growth medium) to remove non-adherent cells, preserving the hydrated extracellular polymeric substance (EPS) matrix.
- For correlated studies, use substrates that are optically transparent for subsequent confocal imaging.
Cantilever Selection and Calibration:
- Selection: Use soft, cantilevers with nominal spring constants of 0.01 - 0.1 N/m to minimize indentation force and avoid sample damage [53].
- Calibration: Precisely calibrate the spring constant of the chosen cantilever using the thermal tune method. Calibrate the optical lever sensitivity on a hard, clean surface (e.g., sapphire) in the same liquid used for imaging.
AFM Setup and Hydration:
- Mount the sample in a liquid cell and ensure it is fully submerged in the appropriate buffer to maintain physiological conditions.
- Carefully mount and align the calibrated cantilever.
- Allow the system to thermally equilibrate for at least 20-30 minutes to minimize thermal drift.
Parameter Optimization for Gentle Imaging:
- For Intermittent Contact Mode: Set a low free oscillation amplitude (1-5 nm) and use a low setpoint (typically >85% of the free amplitude) to ensure the gentlest possible tapping [53].
- For Force Volume Mode: Use a sinusoidal z-modulation to avoid artifacts associated with triangular waveforms. Use a slow ramp speed and a maximum applied force of < 1 nN to stay within the elastic response range of the biofilm [35] [11].
- For Parametric Modes (e.g., Bimodal): Excitate the first two eigenmodes. Use low amplitudes for both modes and adjust the phase shift of the second mode to maximize material contrast while maintaining stable, non-destructive imaging [35].
Data Acquisition and Validation:
- Acquire topography and corresponding property maps (e.g., DMT modulus, adhesion, dissipation).
- Validate sample integrity by comparing pre- and post-scan confocal microscopy images, checking for signs of cell rupture or matrix disruption.
Protocol 2: Correlative AFM-Confocal Workflow for Biofilm Analysis
This workflow integrates AFM nanomechanics with confocal structural data.
- Confocal Pre-Imaging: Acquire a baseline confocal stack of the hydrated biofilm. Use fluorescent stains for cells (e.g., SYTO 9) and EPS (e.g., Con A conjugate) to visualize the 3D structure.
- AFM Nanomechanical Mapping: Transfer the sample to the AFM without allowing it to dehydrate and perform the gentle scanning as detailed in Protocol 1.
- Confocal Post-Imaging: Return the sample to the confocal microscope and re-image the exact same location using fiduciary markers for registration.
- Data Correlation: Co-register the AFM nanomechanical maps (e.g., stiffness) with the confocal channels (e.g., cell location, EPS matrix) using image analysis software (e.g., ImageJ, commercial correlative platforms).
Visualization: Decision Workflow for Gentle AFM Mode Selection
The following diagram outlines a logical pathway for selecting the most appropriate gentle AFM scanning mode based on primary research objectives and sample characteristics.
The Scientist's Toolkit: Essential Reagents and Materials
Successful and reproducible gentle AFM imaging relies on a set of key materials and reagents.
Table 3: Key Research Reagent Solutions for Hydrated Biofilm AFM
| Item | Function/Role | Example Specifications/Notes |
|---|---|---|
| Soft Cantilevers | Transducer for force interaction; soft springs (0.01-0.5 N/m) prevent sample damage. | Silicon nitride probes; triangular or arrow-shaped for stability in liquid. |
| Physiological Buffer | Maintains sample hydration and native state during imaging. | Phosphate Buffered Saline (PBS) or specific bacterial growth medium. |
| Liquid Cell | Enclosed environment to submerge the sample and cantilever in fluid. | Must be compatible with the AFM instrument and sample substrate. |
| Fluorescent Stains | For correlative confocal microscopy; labels cells and EPS components. | SYTO 9 (nucleic acids), Concanavalin A conjugates (polysaccharides). |
| Image Co-registration Software | Aligns and correlates AFM and confocal microscopy datasets. | Open-source (ImageJ) or commercial correlative microscopy platforms. |
| Calibration Reference | For accurate cantilever spring constant and deflection sensitivity calibration. | A clean, rigid sample such as sapphire or a freshly cleaved mica surface. |
The mitigation of sample damage during AFM scanning of hydrated biofilms is not a one-size-fits-all endeavor but a deliberate process of selecting the right tool and methodology. As demonstrated, Intermittent Contact Mode offers the gentlest approach for general topographical imaging, while Force Volume and Nanomechanical Imaging modes provide unparalleled quantitative property data at different speed and resolution trade-offs. The emerging technique of High-Speed AFM is uniquely capable of capturing dynamic processes. Critically, maintaining full hydration, as evidenced in bone tissue studies, is paramount for preserving native structure [54]. By adhering to detailed experimental protocols and employing a correlative workflow with confocal microscopy, researchers can reliably generate high-fidelity nanomechanical data that truly reflects the functional architecture of biofilms, thereby accelerating discovery in drug development and microbial science.
Addressing Probe Contamination in Complex EPS Matrices
Atomic Force Microscopy (AFM) has emerged as a powerful tool for investigating the structural and mechanical properties of bacterial biofilms, particularly their complex extracellular polymeric substance (EPS) matrices. These matrices comprise polysaccharides, proteins, nucleic acids, and lipids that form a protective, three-dimensional scaffold for microbial communities [55] [56]. While AFM provides unprecedented nanoscale resolution for biofilm characterization, the sticky, heterogeneous nature of EPS presents significant challenges for probe contamination, which can compromise data quality and instrument performance. This guide systematically evaluates AFM performance against alternative biofilm analysis techniques, with particular emphasis on experimental approaches that mitigate probe contamination while correlating nanomechanical data with complementary confocal imaging.
Table 1: Comparative Analysis of Biofilm Characterization Techniques
| Technique | Resolution | Probe Contamination Risk | Sample Preparation | Key Advantages | Principal Limitations |
|---|---|---|---|---|---|
| Atomic Force Microscopy (AFM) | 0.5-1 nm lateral, 0.1-0.2 nm axial [57] | High in EPS matrices | Minimal; can image in physiological liquids [7] [57] | Nanomechanical mapping under physiological conditions | Small imaging area (<100 μm²); slow scanning [7] |
| Confocal Laser Scanning Microscopy (CLSM) | 200-300 nm [57] | Not applicable | Fluorescent staining required [55] | 3D visualization of live biofilms; chemical specificity | Diffraction-limited resolution; photobleaching |
| Scanning Electron Microscopy (SEM) | ~1-10 nm | Not applicable | Dehydration and metal coating required [7] | High-resolution surface topography | Non-physiological conditions; potential artifacts |
| Fourier-Transform Infrared Spectroscopy (FTIR) | ~1-10 μm (spatial) [55] | Not applicable | Minimal for transmission mode | Chemical composition analysis | Limited spatial resolution; water interference |
The Probe Contamination Challenge in EPS
The EPS matrix represents a particularly challenging environment for AFM probes due to its chemical complexity and adhesive properties. The matrix evolves structurally and chemically during biofilm maturation, with studies showing increasing lipid content in mature biofilms that enhances resilience and potentially increases adhesion to AFM tips [55]. During scanning, AFM probes can accumulate EPS components, leading to damped oscillation amplitudes in tapping mode, reduced image quality, and altered force measurements that no longer accurately represent true sample properties.
The contamination risk is particularly pronounced when investigating mature, dense biofilms where the dense network of polymeric substances creates a highly adhesive scanning environment. This necessitates specific experimental strategies to distinguish genuine sample properties from artifact data resulting from contaminated probes.
Experimental Approaches for Contamination Mitigation
Probe Selection and Functionalization
Probe specifications for biofilm imaging should prioritize sharp tips (high aspect ratio) with appropriate surface chemistry to minimize adhesion. Silicon nitride probes with spring constants of approximately 0.1 N/m are suitable for soft biofilm samples in liquid environments [57]. For higher-resolution imaging of substructures like flagella (20-50 nm in height), sharper silicon tips are preferable but require more careful contamination monitoring [7].
Surface functionalization strategies can reduce nonspecific adhesion:
- Hydrophilic coatings (PEG) create a hydration barrier that resists EPS adhesion
- Self-assembled monolayers with controlled surface energy can be tuned for specific EPS components
- Chemical passivation using bovine serum albumin (BSA) or other blocking proteins can occupy adhesive sites on the probe surface
Operational Methodologies
Large-area automated AFM represents a significant advancement for biofilm characterization, combining machine learning with automated scanning to capture high-resolution images over millimeter-scale areas [7]. This approach minimizes human intervention and enables the identification of representative scanning areas with lower contamination risk.
HS-AFM (High-Speed AFM) reduces tip-sample interaction time, potentially decreasing contamination buildup while enabling observation of dynamic processes [57]. However, it currently limits scanning areas to approximately 1 μm², restricting larger-scale architectural analysis.
Multimodal integration with confocal microscopy provides a correlation framework where AFM can target specific, well-characterized regions identified via confocal imaging, reducing unnecessary scanning through high-contamination areas [7].
Probe Cleaning and Validation Protocols
In-situ cleaning methods include:
- UV-ozone treatment between scans to oxidize organic contaminants
- Solvent rinsing with appropriate buffers (requires recalibration)
- Plasma cleaning for more thorough decontamination (requires probe removal)
Contamination monitoring should include regular verification scans on reference samples with known topography and mechanical properties. Automated probe conditioning approaches using machine learning algorithms can maintain consistent tip geometry throughout extended experiments [7].
Table 2: Research Reagent Solutions for AFM Biofilm Studies
| Reagent/Material | Function | Application Notes |
|---|---|---|
| Silicon nitride probes with reflective coating | Biofilm imaging and force spectroscopy | Optimal spring constant ~0.1 N/m for live cell imaging [57] |
| PFOTS-treated glass surfaces | Hydrophobic substrate for controlled biofilm growth | Used in Pantoea sp. YR343 studies to observe cellular orientation patterns [7] |
| SYTO9/PI fluorescent stains | Viability assessment via CLSM correlation | Enables direct correlation of nanomechanical properties with cell viability [58] |
| Modified C2+ intravascular lithotripsy catheter | Shockwave biofilm disruption for comparative studies | 120 pulses at 2 Hz parameters effective for EPS disruption [58] |
| Crystal violet solution | Biofilm biomass quantification | Standard 1% solution, 20-minute staining at 37°C [58] |
Correlative AFM-Confocal Workflow for Biofilm Analysis
The integration of AFM nanomechanics with confocal biofilm imaging creates a powerful correlative approach that contextualizes mechanical properties within architectural and compositional frameworks. The following experimental workflow optimizes this correlation while minimizing probe contamination:
Workflow for Correlative AFM-Confocal Biofilm Analysis
Protocol Details
Sample Preparation: Grow biofilms on optically transparent substrates (glass coverslips) compatible with both techniques. For Pseudomonas aeruginosa and Escherichia coli biofilms, standard growth periods of 24-72 hours produce mature architectures with developed EPS matrices [55].
Confocal Imaging: Acquire 3D structural data using appropriate fluorescent markers (SYTO9 for live cells, ConA for polysaccharides). CLSM reveals biofilm thickness, cell distribution, and spatial arrangement within the EPS matrix [55].
Region Selection: Identify representative areas with minimal debris or extreme thickness that might promote probe contamination. Machine learning algorithms can assist in selecting optimal scanning regions [7].
AFM Nanomechanical Mapping: Employ tapping mode in liquid to minimize shear forces. Collect force-volume maps at multiple locations within the confocal-identified region to capture mechanical heterogeneity.
Data Correlation: Overlay AFM-derived mechanical properties (Young's modulus, adhesion) with confocal structural data to establish structure-property relationships in the biofilm EPS.
Probe Validation: Regular verification scans on reference polydimethylsiloxane (PDMS) samples with known mechanical properties monitor contamination status throughout the experiment.
Comparative Performance Data
Recent studies quantitatively demonstrate AFM capabilities in biofilm characterization alongside contamination challenges:
Structural resolution: AFM successfully identified flagellar structures approximately 20-50 nm in height and extending tens of micrometers across surfaces in Pantoea sp. YR343 biofilms, a resolution unmatched by optical techniques [7].
Mechanical characterization: Force spectroscopy revealed significant mechanical heterogeneity within Pseudomonas aeruginosa biofilms, with Young's modulus values varying from 10 kPa to 2 MPa across different EPS regions.
Contamination impact: Studies reported 40-60% reduction in measured adhesion forces and 25-35% increase in apparent stiffness values when using contaminated probes on standardized hydrogel samples mimicking biofilm mechanical properties.
Large-area analysis: Automated large-area AFM achieved millimeter-scale imaging while maintaining 5-10 nm resolution, representing a 1000-fold increase in scan area compared to conventional AFM while managing contamination through automated tip conditioning protocols [7].
Future Perspectives
Emerging approaches to address probe contamination in complex EPS matrices include:
- AI-driven probe conditioning: Machine learning algorithms that automatically maintain probe integrity through adaptive cleaning cycles [7]
- Advanced tip functionalization: Non-fouling polymer brushes and slippery liquid-infused surfaces that resist EPS adhesion
- Multi-modal integration: Combined AFM-optical systems that enable real-time contamination monitoring during scanning
- Standardized reference materials: Biofilm-mimicking hydrogels with certified mechanical properties for contamination assessment
These developments, coupled with the growing integration of AFM with complementary techniques like confocal microscopy, will continue to enhance the reliability and applicability of AFM for investigating the complex biomechanical properties of biofilms and their EPS matrices.
Solving Image Registration Challenges Between AFM and CLSM Datasets
In the study of complex biological systems like biofilms, researchers increasingly rely on correlative microscopy to combine the high-resolution nanomechanical data from Atomic Force Microscopy (AFM) with the detailed molecular and structural information from Confocal Laser Scanning Microscopy (CLSM). This multi-modal approach provides a comprehensive picture of biofilm architecture, function, and response to environmental stressors [59]. However, the integration of these disparate datasets presents significant image registration challenges arising from fundamental differences in how each technique acquires data, their respective resolutions, fields of view, and the types of information they prioritize.
The registration between AFM and CLSM is not merely a technical exercise but a fundamental requirement for advancing our understanding of structure-function relationships in microbial systems. AFM provides exceptional nanoscale topographical imaging and quantitative mechanical mapping (including elasticity, adhesion, and viscosity) under physiologically relevant conditions, but offers limited information on internal structures or molecular composition [4] [5]. CLSM, meanwhile, excels at optical sectioning and 3D reconstruction of fluorescently tagged components within a sample, but suffers from diffraction-limited resolution and cannot directly measure mechanical properties [60]. When successfully correlated, these techniques enable researchers to precisely map mechanical properties to specific structural features and molecular markers within biofilms, opening new avenues for understanding antibiotic resistance, host-pathogen interactions, and environmental adaptation [59] [61].
Fundamental Technical Challenges in AFM-CLSM Registration
Dimensional and Resolution Mismatches
The integration of AFM and CLSM data faces inherent technical hurdles rooted in their operational principles. The most immediate challenge is the scale discrepancy between the instruments. Conventional AFM has a limited scan range (typically <100 µm) determined by piezoelectric actuator constraints, while CLSM can readily image millimeter-scale areas [7]. This creates a field-of-view mismatch that complicates the registration of local nanomechanical properties to larger biofilm architectures.
Furthermore, the resolution characteristics of each technique differ substantially. AFM provides exceptional lateral resolution at the nanometer scale, allowing visualization of fine structures like bacterial flagella (20-50 nm in height) and surface proteins [7]. CLSM, while excellent for its purpose, is diffraction-limited, with best-case lateral resolution of approximately 200 nm and axial resolution around 600 nm [60]. This resolution gap means that features clearly resolved by AFM may be near or below the detection limit of CLSM, creating ambiguity in feature matching during registration.
Temporal and Spatial Acquisition Differences
The acquisition methodology presents another layer of complexity. AFM is a surface scanning technique that constructs images point-by-point through physical probe interaction, making it relatively slow and potentially disruptive to delicate biological samples [59]. CLSM, particularly in spinning disk implementations, can capture dynamic processes at much higher speeds but requires fluorescent labeling that may alter native biological activity [60] [62].
The dimensional orientation of datasets also differs. AFM primarily captures topographical surface data, while CLSM collects optical sections through a volume, creating a challenge in aligning 2.5D AFM data with true 3D CLSM reconstructions. Additionally, each technique has unique artifact generation: AFM may produce tip-convolution effects and sample deformation, while CLSM contends with photobleaching, scattering, and refractive index mismatches [59] [60].
Comparative Analysis of Registration Methodologies
Researchers have developed several strategic approaches to overcome these registration challenges, each with distinct advantages and limitations. The table below provides a comparative analysis of the primary methodologies employed for AFM-CLSM correlation.
Table 1: Comparison of AFM-CLSM Image Registration Methodologies
| Methodology | Core Principle | Best Application Context | Key Advantages | Inherent Limitations |
|---|---|---|---|---|
| Fiducial Marker-Based Registration | Uses physical reference points visible to both techniques | Live-cell imaging; dynamic processes | High precision alignment; quantitative validation capability | Potential interference with native biology; marker size constraints |
| Large-Area AFM with Automated Stitching | Machine learning-assisted stitching of multiple AFM scans | Biofilm architecture studies; heterogeneous samples | Bridges scale gap; preserves high-resolution context | Computational intensity; requires specialized instrumentation |
| Simultaneous Integrated Imaging (Conpokal) | Physical instrument integration for concurrent data acquisition | Real-time mechanobiological responses; nanoscale dynamics | Eliminates temporal delays; inherent spatial registration | Instrument complexity; potential optical interference |
| Feature-Based Algorithmic Registration | Computational matching of recognizable structural features | Fixed samples; well-defined cellular structures | No foreign markers needed; utilizes native sample features | Feature recognition challenges across resolution scales |
Fiducial Marker-Based Registration
The use of fiducial markers remains one of the most reliable approaches for precise AFM-CLSM registration. This method involves incorporating reference structures with distinct signatures in both modalities, such as fluorescent microspheres with well-defined topographical profiles or patterned substrates with recognizable geometries.
In practice, quantum dot nanocrystals have proven particularly valuable as they provide strong fluorescence signals for CLSM detection while maintaining structural integrity for AFM imaging [63]. The application of ultra-pure methanol-free formaldehyde fixation has been shown to preserve both fluorescence properties and nanoscale ultrastructure, which is crucial for maintaining registration accuracy [63]. This approach enabled researchers to make the novel discovery that filopodia possess a "quilted" surface structure and to directly visualize moesin linkages between transmembrane proteins and the cytoskeleton [63].
Large-Area AFM with Automated Stitching
For biofilm research where architectural context is essential, large-area AFM approaches combined with computational stitching address the critical field-of-view limitation. Recent advancements have demonstrated automated AFM systems capable of scanning millimeter-scale areas while maintaining nanometer resolution, effectively bridging the scale gap between cellular and community-level organization [7].
The integration of machine learning algorithms has been transformative for this approach, enabling intelligent site selection, seamless image stitching with minimal overlap, and automated analysis of stitched datasets [7]. In studying Pantoea sp. YR343 biofilm formation, this methodology revealed previously obscured spatial heterogeneity, including a preferred cellular orientation forming distinctive honeycomb patterns and the coordinated role of flagella in community assembly beyond initial attachment [7]. The computational workflow for this approach can be visualized as follows:
Diagram 1: Large-Area AFM-CLSM Registration Workflow
Simultaneous Integrated Imaging (Conpokal)
The most technically advanced approach involves fully integrated AFM-CLSM systems that enable simultaneous data acquisition. This methodology, sometimes called "Conpokal," provides inherent spatial and temporal registration by design [62]. These systems permit researchers to directly correlate mechanical properties with dynamic cellular processes in real-time, essentially allowing scientists to "see what they are probing" [62].
The implementation of AFM quantitative imaging (QI) mode coupled with simultaneous LSCM has been particularly successful for live-cell investigations. This combination produces multiplexed data on cell morphology, mechanics, surface adhesion, and ultrastructure while tracking multiple fluorescently tagged macromolecules in real-time [61]. Studies employing this methodology have revealed how xenobiotics like 2,4-dichlorophenoxyacetic acid induce oxidative stress and disrupt cellular structures across bacterial, fungal, and human cell models [61].
Experimental Protocols for Robust AFM-CLSM Registration
Sample Preparation for Correlative Imaging
Proper sample preparation is foundational to successful AFM-CLSM registration. The protocol must simultaneously optimize conditions for both fluorescence preservation and nanoscale structural integrity. Based on successful methodologies reported in the literature, the following procedure has demonstrated efficacy:
- Fixation Approach: Use ultra-pure methanol-free formaldehyde (e.g., 4% in physiological buffer) for 20-30 minutes at room temperature. This formulation maximizes fluorescence response while maintaining epithelial cell ultrastructure essential for high-quality AFM imaging [63].
- Substrate Selection: Employ glass-bottomed Petri dishes coated with poly-L-lysine or similar adhesives to ensure firm attachment during AFM scanning while maintaining optical clarity for CLSM [62]. For specialized biofilm studies, chemically modified surfaces (e.g., PFOTS-treated glass) provide controlled attachment conditions [7].
- Fluorescent Labeling: Incorporate quantum dot nanocrystals or bright fluorescent proteins with minimal steric hindrance. For live-cell imaging, CellROX dyes for reactive oxygen species detection combined with FtsZ-GFP for structural markers have proven effective [61].
- Immobilization Verification: Confirm sample stability through preliminary AFM scanning in liquid using force mapping modes with minimal applied force (typically 0.1-0.5 nN) to prevent sample displacement [61].
Data Acquisition Parameters
Consistent acquisition parameters are critical for reproducible registration across instruments and experimental sessions:
Table 2: Optimal Acquisition Parameters for AFM-CLSM Correlation
| Parameter | AFM Settings | CLSM Settings | Rationale |
|---|---|---|---|
| Scan Size | 10×10 µm to 100×100 µm (stitched if larger) | Match to AFM scan area | Ensures direct spatial correspondence |
| Resolution | 512×512 pixels or higher | 512×512 pixels or higher | Maintains detail for feature matching |
| Scan Rate | 0.5-1.0 Hz | 0.5-1.0 fps (for time series) | Minimizes temporal discrepancy |
| Laser Wavelength | N/A | 488 nm (GFP), 561 nm (RFP), 640 nm (far-red) | Standard fluorophore excitation |
| Force Settings | 0.1-0.5 nN setpoint, spherical probes (R = 5,000 nm) | N/A | Prevents sample damage, ensures reliable contact mechanics |
| Optical Sectioning | N/A | 0.5-1.0 µm z-steps | Adequate 3D reconstruction without excessive photobleaching |
Computational Registration Workflow
After data acquisition, computational methods refine the alignment and enable quantitative correlation:
- Preprocessing: Apply flat-field correction to CLSM images and leveling to AFM height images to remove background trends and scanner bow.
- Feature Detection: Identify common structures (cell boundaries, distinctive clusters, or intentionally introduced fiducials) in both modalities using edge detection algorithms or machine learning-based segmentation [7].
- Transformation Calculation: Compute optimal affine transformation (rotation, translation, scaling) to align AFM and CLSM datasets using point-based registration or intensity-based correlation algorithms.
- Validation: Quantify registration accuracy by measuring alignment error of control points not used in transformation calculation, with successful registration typically achieving sub-micrometer accuracy.
The following diagram illustrates the integrated experimental workflow that incorporates these protocols:
Diagram 2: Integrated AFM-CLSM Experimental Workflow
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful AFM-CLSM correlation requires careful selection of specialized materials and reagents. The following table catalogues essential solutions for researchers designing correlative microscopy studies of biofilms and cellular systems.
Table 3: Essential Research Reagent Solutions for AFM-CLSM Correlation
| Reagent/Material | Function | Application Notes | Key References |
|---|---|---|---|
| Ultra-pure Methanol-free Formaldehyde | Chemical fixation | Preserves fluorescence and ultrastructure; superior to glutaraldehyde for correlative work | [63] |
| Quantum Dot Nanocrystals | Fluorescent fiducial markers | Photostable reference points for registration; various sizes available | [63] |
| Poly-L-Lysine Coated Glass-bottom Dishes | Sample substrate | Provides firm attachment for AFM while maintaining optical clarity for CLSM | [62] |
| Cell-Tak Adhesive | Sample immobilization | Effective for challenging samples like bacteria; maintains viability | [61] |
| Borosilicate Glass Spherical Probes (R = 5,000 nm) | AFM cantilevers | Ideal for soft biological samples; reduces sample damage | [64] |
| CellROX Oxidative Stress Reagents | Fluorescent stress indicators | Measure ROS production in live cells during mechanical characterization | [61] |
| FtsZ-GFP and Tubulin2-GFP Plasmids | Structural fluorescent markers | Visualize cytoskeletal elements during nanomechanical profiling | [61] |
| PFOTS-treated Glass Surfaces | Controlled attachment substrates | Standardize bacterial adhesion for reproducible biofilm studies | [7] |
The registration of AFM and CLSM datasets remains challenging but increasingly achievable through methodological advances in sample preparation, instrumentation, and computational analysis. The most promising developments include the continued refinement of fully integrated AFM-CLSM systems that eliminate temporal discrepancies, the application of machine learning for automated large-area correlation, and the development of novel fiducial markers that provide unambiguous registration points without interfering with native biology.
As these technologies mature, researchers will be better equipped to tackle fundamental questions in biofilm mechanobiology, from how mechanical properties influence antibiotic resistance to how nanoscale surface interactions determine community architecture. The systematic comparison of registration methodologies presented in this guide provides a foundation for selecting appropriate strategies based on specific research requirements, sample characteristics, and available instrumentation. Through continued methodological refinement, the correlation of nanomechanical and optical datasets will undoubtedly yield new insights into the intricate world of microbial communities and their interactions with surfaces, antibiotics, and host systems.
Leveraging Automation and Machine Learning for Large-Area Analysis and Segmentation
Biofilms are complex, heterogeneous microbial communities whose remarkable resilience and function are dictated by their intricate spatial architecture and mechanical properties. Understanding these structures requires techniques that can capture data across multiple scales, from single macromolecules to entire communities. Atomic force microscopy (AFM) provides unparalleled nanoscale resolution of surface ultrastructure and nanomechanical properties, while confocal laser scanning microscopy (CLSM) offers a window into the three-dimensional internal organization of live biofilms. The correlation of these datasets is crucial for a comprehensive picture of biofilm behavior [61] [65]. However, traditional AFM is limited by its small imaging area (typically <100 µm), restricted by piezoelectric actuator constraints, making it difficult to capture the full spatial complexity of millimeter-scale biofilm structures and raising questions about data representativeness [7]. This gap between nanoscale cellular features and macroscale community organization has historically hindered a complete understanding of biofilm assembly and function.
The integration of automation and machine learning (ML) is now revolutionizing this field, enabling large-area analysis and segmentation that bridges this scale gap. These technologies transform AFM from a manual, single-image technique into a high-throughput platform capable of quantifying heterogeneity across entire biofilm samples. Concurrently, ML algorithms are automating the complex segmentation and analysis of CLSM data, moving beyond qualitative assessments to provide robust, quantitative metrics of biofilm viability and architecture. This guide compares the emerging methodologies that are setting new standards for performance in correlative AFM-confocal biofilm research, providing researchers with the data needed to select the optimal approach for their specific applications.
Comparative Analysis of Emerging Technologies
The following technologies represent the forefront of automated and ML-powered solutions for large-area biofilm characterization. They are evaluated based on their core methodology, key performance metrics, and applicability to correlated AFM-confocal studies.
Table 1: Technology Comparison for Large-Area Biofilm Analysis
| Technology / Approach | Core Methodology | Reported Performance Metrics | Key Advantages | Primary Limitations |
|---|---|---|---|---|
| Automated Large-Area AFM [7] | Automated tiling of high-resolution AFM scans with ML-based stitching and cell detection. | • Area: Millimeter-scale• Resolution: Nanoscale (visualizes ~20-50 nm flagella)• Throughput: Enables multiday experiments without supervision | • Captures preferred cellular orientation & emergent patterns (e.g., honeycomb)• Directly links nanoscale features to macroscale organization | • Requires rigid, dried samples for optimal high-resolution imaging |
| AI-Empowered CLSM Analysis [10] | Deep learning (e.g., CNN) for automated segmentation, classification, and 3D quantification of CLSM images. | • Analysis Speed: High-throughput, automated• Accuracy: Enhanced resolution and artifact reduction• Output: Quantifies spatial heterogeneity, biovolume, and cell viability | • Analyzes complex 3D structures under physiological conditions• Superior for soft, hydrated biofilms and viability staining (e.g., Live/Dead) | • Indirect measurement of surface mechanics and properties |
| Correlative AFM-QI-LSCM [61] | Simultaneous physical integration of AFM in Quantitative Imaging (QI) mode with Live Cell Confocal Microscopy. | • AFM Resolution: nm-scale (topography), pN-scale (mechanics)• Data: Multiplexed nanomechanics & real-time fluorescence• Application: Live cell tracking under stress | • True real-time correlation of surface mechanics and internal molecular events• Probes biomechanics and physiology in native hydrated state | • Limited scan area compared to automated large-area AFM• Complex instrument operation |
Table 2: Quantitative Data Output from Featured Studies
| Study | Cell Type / Biofilm | Key Quantitative Findings | Source |
|---|---|---|---|
| Automated Large-Area AFM | Pantoea sp. YR343 | Revealed a preferred cellular orientation forming a distinctive honeycomb pattern; identified flagellar structures (~20-50 nm height) bridging gaps between cells. | [7] |
| AI-Empowered CLSM | Multi-species oral biofilm | Automated analysis showed a coefficient of variation (CV) of 4.24-11.5%, significantly lower than the 17.0-78.1% CV for traditional CFU counting methods. | [42] |
| Correlative AFM-QI-LSCM | Candida albicans (Fungal) | Exposure to 8 mM 2,4-D caused a significant reduction in surface roughness and a two-fold increase in both surface adhesion and elasticity (Young's Modulus). | [61] |
Experimental Protocols for Key Methodologies
Protocol: Automated Large-Area AFM for Biofilm Assembly Analysis
This protocol is adapted from the workflow that enabled millimeter-scale analysis of Pantoea sp. YR343 biofilms [7].
Sample Preparation:
- Grow biofilms on adhesion-promoting substrates (e.g., PFOTS-treated glass coverslips).
- At desired time points, gently rinse the coverslip to remove unattached planktonic cells.
- Critical Step: Air-dry the sample prior to imaging. This protocol requires dried samples to achieve high-resolution nanoscale imaging of features like flagella.
Instrument Setup and Automated Scanning:
- Mount the sample on the AFM stage.
- Define the large-area (mm-scale) region of interest using the microscope's software.
- Initiate the automated tiling routine. The system will sequentially scan multiple adjacent tiles with minimal user intervention.
Machine Learning-Enhanced Data Processing:
- Image Stitching: Use ML-driven algorithms to seamlessly merge the individual high-resolution tiles into a single, large-area map, compensating for stage drift and imperfections.
- Cell Detection and Classification: Apply trained ML models to automatically identify individual cells within the stitched image, extract parameters like cell count, confluency, shape, and orientation.
Data Validation: Manually verify the output of the automated analysis against a subset of the original data to ensure algorithm accuracy.
Protocol: Automated Viability Analysis of Biofilms via CLSM
This protocol leverages an open-source tool developed for robust quantification of biofilm viability from CLSM z-stacks, validated against traditional microbiology [42].
Sample Staining and Imaging:
- Staining: Treat hydrated, live biofilms with a viability stain, such as the FilmTracer LIVE/DEAD BacLight kit (SYTO 9 and propidium iodide).
- Image Acquisition: Collect z-stack images of the biofilms using a Confocal Laser Scanning Microscope (CLSM). Set appropriate laser wavelengths and detection bands for the green (live cells) and red (dead cells) fluorescence channels.
Image Pre-processing (in FIJI/ImageJ):
- Import the z-stack images.
- Apply a median filter (e.g., 2-pixel radius) to reduce noise while preserving edges.
- Split the channels for independent analysis of the green and red signals.
Automated Segmentation and Thresholding:
- For each channel, apply an automated thresholding algorithm (e.g., Otsu, Li, or Triangle method). This step objectively differentiates signal from background, eliminating user bias.
- Perform morphological operations (e.g., "Fill Holes," "Watershed") to refine the segmentation and separate touching cells.
Quantification and Output:
- Use the "Analyze Particles" function to quantify the area, volume, and pixel intensity for each channel.
- Calculate the percentage of live and dead cells based on the segmented areas. The analysis macro outputs quantitative data on biomass and viability, with significantly lower coefficient of variation than manual CFU counting [42].
Diagram Title: Automated CLSM Biofilm Viability Analysis Workflow
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful execution of the described protocols relies on a set of key materials and software solutions.
Table 3: Essential Research Reagents and Solutions
| Item Name | Function / Application | Critical Notes |
|---|---|---|
| PFOTS-treated Glass | Creates a highly hydrophobic surface to promote robust bacterial adhesion for stable AFM imaging. | Crucial for immobilizing motile cells like Pantoea sp. for large-area AFM [7]. |
| FilmTracer LIVE/DEAD Biofilm Viability Kit | Two-color fluorescent stain (SYTO 9 & Propidium Iodide) to distinguish live vs. dead cells based on membrane integrity in CLSM. | Automated analysis of separate channels is essential for objective quantitation, avoiding misleading color superposition [42]. |
| FIJI / ImageJ Software | Open-source platform for image analysis. Serves as the foundation for running custom macros for automated biofilm quantification. | The "Biofilm Viability Checker" macro is built here, making analysis accessible to non-specialists [42]. |
| Cell-Tak | Bioadhesive used to immobilize cells (e.g., E. coli) onto substrates for correlative AFM-confocal experiments with live cells in liquid. | Note: Effectiveness may diminish after multiple cell divisions, allowing daughter cells to detach [61]. |
| Polydimethylsiloxane (PDMS) Stamps | Micro-patterned stamps used for the mechanical immobilization of spherical microbial cells for single-cell AFM analysis in liquid. | Provides secure immobilization without chemical fixation, preserving native physiological and nanomechanical properties [65]. |
Diagram Title: Logical Flow from Problem to Solution in Biofilm Analysis
The integration of automation and machine learning with AFM and confocal microscopy is fundamentally advancing biofilm research. As the comparative data shows, the choice between cutting-edge approaches involves a strategic trade-off: automated large-area AFM provides unparalleled nanoscale structural and mechanical data across millimeter scales but typically on dried samples, while AI-empowered CLSM analysis offers powerful, high-throughput 3D volumetric and viability data from live, hydrated biofilms. The fully correlative AFM-QI-LSCM platform stands apart for its unique ability to probe nanomechanics and internal molecular events in live cells simultaneously, albeit over smaller areas.
For researchers and drug development professionals, the optimal path forward lies in selecting the technology that best aligns with their primary research question. The ongoing refinement of these tools, particularly through enhanced AI and multimodal data fusion, promises not only to deepen our fundamental understanding of biofilm heterogeneity and resilience but also to accelerate the discovery and efficacy testing of novel anti-biofilm therapeutic strategies.
Benchmarking Performance: How Correlative AFM-Confocal Compares to Traditional Methods
Biofilms are complex, three-dimensional microbial communities that pose significant challenges in medical, industrial, and environmental contexts due to their resilience against antibiotics and disinfectants [66]. For decades, research and diagnostics have relied heavily on two fundamental methods for biofilm quantification: crystal violet (CV) staining for total biomass assessment and colony forming unit (CFU) enumeration for viable cell counts. While these methods have provided valuable foundational data, a growing body of evidence reveals critical limitations that constrain their effectiveness, particularly for sophisticated applications like correlating nanomechanical properties with architectural features. The crystal violet method, despite its widespread utilization, is not without flaws, especially when assessing multi-species biofilms where it may provide significant bias [67]. Similarly, CFU counts can underestimate true cell numbers due to the presence of viable but non-culturable (VNBC) cells and fail to capture the spatial organization critical to biofilm function [68]. This article examines the limitations of these traditional methods and explores advanced techniques that provide more comprehensive biofilm characterization, with particular focus on applications for correlating atomic force microscopy (AFM) nanomechanics with confocal biofilm imaging research.
Critical Limitations of Traditional Biofilm Assessment Methods
Crystal Violet Staining: Beyond Biomass Quantification
Crystal violet staining serves as an indirect measurement of biofilm formation by binding to negatively charged surface molecules and polysaccharides in the extracellular matrix. However, this method provides no information on cell viability, metabolic activity, or three-dimensional organization within the biofilm [66]. Recent research specifically demonstrates that CV staining fails to properly quantify in vitro multi-species biofilms, such as those composed of common BV-associated bacteria (Gardnerella vaginalis, Fannyhessea vaginae, and Prevotella bivia), where it cannot distinguish possible synergism or antagonism between individual species [67]. When comparing CV method with total CFU and fluorescence microscopy cell count methods for single-species biofilms, the relationship between biofilm biomass, total number of cells, and total cultivable cells varied significantly between each tested method and also changed with incubation time [67]. This fundamental limitation makes CV staining inadequate for pathogenicity research or antimicrobial efficacy testing where viability and community interactions are crucial parameters.
CFU Enumeration: The Viability Gap
The CFU technique quantifies viable, culturable cells but fails to account for several biologically significant populations within biofilms. These limitations include:
- Underestimation of true cell numbers due to bacterial clumping and the presence of viable but non-culturable (VBNC) cells [66] [68]
- Inability to detect population heterogeneity within biofilms, including metabolically dormant persister cells that contribute significantly to antibiotic tolerance [68]
- Time-intensive processes requiring 24-72 hours for results, preventing real-time assessment [66]
- Culture condition dependence that may selectively favor certain species over others in polymicrobial biofilms [67]
These limitations are particularly problematic in clinical diagnostics and treatment evaluation, where accurate assessment of all viable bacterial populations is essential for effective therapeutic interventions.
Table 1: Fundamental Limitations of Traditional Biofilm Assessment Methods
| Parameter | Crystal Violet Staining | CFU Enumeration |
|---|---|---|
| Viability Information | No distinction between live and dead cells | Only detects culturable cells |
| Spatial Organization | No 3D structural data | No spatial information |
| Matrix Composition | Indirect measurement, non-specific | No matrix information |
| Time Requirements | Rapid (hours) | Slow (24-72 hours) |
| Multi-species Applications | Poor reliability for complex communities | Culture bias affects accuracy |
| Mechanical Properties | No data on biofilm mechanics | No data on biofilm mechanics |
Advanced Methodologies for Comprehensive Biofilm Analysis
Atomic Force Microscopy (AFM) for Nanoscale Structural and Mechanical Characterization
Atomic force microscopy provides unprecedented capabilities for characterizing biofilms at the nanoscale, offering both topographical imaging and quantitative mapping of nanomechanical properties without extensive sample preparation [7]. Recent technological advances have addressed traditional AFM limitations through automated large-area AFM approaches capable of capturing high-resolution images over millimeter-scale areas, enabling researchers to link cellular and subcellular features to functional macroscale organization [7].
Key Applications of AFM in Biofilm Research:
- Nanomechanical mapping: Measurement of stiffness, adhesion, and viscoelastic properties of individual cells and biofilm matrix components [7]
- High-resolution topography: Visualization of structural features surpassing the resolution of optical or electron beam-based microscopy, including membrane protrusions, surface proteins, and cell wall ridges [7]
- Real-time monitoring: Capability to image under physiological conditions, enabling observation of dynamic structural changes [7]
- Correlative imaging: Integration with other microscopic techniques to connect structural features with biological functions [69]
The application of machine learning and artificial intelligence has further transformed AFM by enhancing data acquisition, control, and analysis, enabling autonomous operation and continuous multi-day experiments without human supervision [7]. For instance, large-area AFM has revealed distinctive honeycomb patterns and preferred cellular orientation during early biofilm formation of Pantoea sp. YR343, along with detailed mapping of flagella interactions measuring ~20-50 nm in height and extending tens of micrometers across surfaces [7].
Flow Cytometry for Rapid Phenotypic Characterization
Flow cytometry offers a powerful high-throughput alternative for bacterial phenotyping and growth phase assessment that overcomes several limitations of traditional methods. A dual-stain procedure utilizing cell-permeable SYTO 60 and membrane-impermeable TOTO-1 dyes enables accurate identification and quantitation of biofilm cells versus planktonic cells [68]. This method specifically identifies biofilm growth phase as double positive for both SYTO 60 and TOTO-1, while normal logarithmic growth phase is largely negative for TOTO-1 staining [68].
Experimental Protocol for Flow Cytometric Biofilm Assessment:
- Prepare bacterial single cell suspension from biofilm cultures
- Stain with SYTO 60 (cell-permeable DNA dye) and TOTO-1 (membrane-impermeable eDNA dye)
- Add polystyrene counting beads for accurate quantitation
- Analyze using flow cytometer with SSC threshold set using buffer-only control
- Identify biofilm populations as dual-positive for both fluorescent markers
This approach has demonstrated specificity through comparison with impaired biofilm-forming strains of P. aeruginosa LasI/RhlI−/− and ∆PfPhage, showing significantly reduced dual-positive populations in these mutants compared to wildtype strains [68]. The method provides substantial advantages over traditional techniques, including rapid processing, high-throughput capability, and ability to detect population-level heterogeneity.
Advanced Microscopy and Spectroscopic Techniques
Correlative approaches that combine multiple imaging modalities provide the most comprehensive understanding of biofilm architecture and function:
- Confocal Laser Scanning Microscopy (CSLM): Provides three-dimensional images of biofilms without disruption, especially when combined with fluorescent staining techniques [66]
- Large-Area Automated AFM: Enables high-resolution imaging over millimeter-scale areas, bridging the gap between nanoscale features and macroscale organization [7]
- Fluorescence Microscopy with Advanced Staining: Uses combination stains to distinguish between live cells, dead cells, and extracellular DNA components [68]
Table 2: Advanced Biofilm Characterization Techniques and Their Applications
| Technique | Resolution | Key Measurable Parameters | Advantages Over Traditional Methods |
|---|---|---|---|
| Atomic Force Microscopy | Nanoscale | Topography, stiffness, adhesion, viscoelasticity | Quantitative nanomechanical data under physiological conditions |
| Flow Cytometry | Single cell | Viability, eDNA content, population heterogeneity | High-throughput, quantitative population analysis |
| Confocal Microscopy | Sub-micron | 3D architecture, spatial organization, chemical gradients | Non-destructive 3D imaging of hydrated biofilms |
| Large-Area AFM | Nanoscale over mm areas | Cellular orientation, appendage interactions, surface adhesion | Links nanoscale features to macroscale organization |
| Advanced Staining Methods | Cellular | Live/dead differentiation, matrix composition | Specific identification of biofilm components |
Experimental Designs for Correlative AFM and Confocal Imaging
Integrated Workflow for Structural and Mechanical Correlation
Establishing correlation between AFM nanomechanics and confocal biofilm imaging requires carefully designed experimental protocols that preserve sample integrity while enabling multi-modal analysis:
Sample Preparation Protocol:
- Grow biofilms on appropriate substrates compatible with both AFM and confocal microscopy (e.g., glass coverslips, titanium coupons)
- For structural assessment: stain with appropriate fluorescent markers (e.g., SYTO 60 for cells, TOTO-1 for eDNA)
- For AFM imaging: gently rinse to remove unattached cells but preserve biofilm integrity
- For mechanical characterization: maintain hydration during AFM analysis to preserve native properties
Correlative Imaging Workflow:
- Initial confocal imaging to identify regions of interest based on architectural features
- AFM nanomechanical mapping of identified regions for stiffness and adhesion measurements
- Large-area AFM scanning to connect local mechanical properties with broader community organization
- Data integration using machine learning approaches to correlate structural and mechanical parameters
This approach has been successfully applied to study bacterial species such as Pantoea sp. YR343, revealing how flagellar coordination contributes to biofilm assembly beyond initial attachment [7].
Experimental Models for Biofilm Research
Choosing appropriate experimental models is crucial for generating clinically relevant data:
- Modified Bone-Like Environment (BLE+): For prosthetic joint infection biofilms, incorporating physiological oxygen concentrations (2.5% O₂) significantly impacts biofilm formation compared to standard anaerobic conditions [70]
- Suspended peg systems: Eliminate interference from sedimented aggregates by driving active bacterial adhesion related specifically to biofilm formation [70]
- Drip Flow Reactors and CDC Biofilm Reactors: Provide reproducible biofilm growth on medical device material surrogates with low variability across replicates [71]
- Multi-species models: Incorporate relevant bacterial interactions, such as the synergy between Gardnerella vaginalis, Fannyhessea vaginae, and Prevotella bivia in BV-associated biofilms [67]
Essential Research Reagent Solutions
Successful implementation of advanced biofilm characterization methods requires specific research reagents and materials:
Table 3: Key Research Reagent Solutions for Advanced Biofilm Studies
| Reagent/Material | Function | Application Examples |
|---|---|---|
| SYTO 60 | Cell-permeable nucleic acid stain | Labels all bacterial cells in flow cytometry and fluorescence microscopy |
| TOTO-1 | Membrane-impermeable DNA stain | Specific detection of extracellular DNA in biofilm matrix |
| Polystyrene counting beads | Quantitative reference standard | Enables accurate cell counting in flow cytometry |
| PFOTS-treated glass | Hydrophobic surface modification | Controls bacterial adhesion for AFM studies |
| BLE+ medium | Physiologically relevant culture medium | Supports biofilm growth in prosthetic joint infection models |
| Titanium coupons/pegs | Medical device material surrogate | Studies of biofilm formation on orthopedic implants |
The limitations of traditional biofilm assessment methods like crystal violet staining and CFU enumeration have become increasingly apparent as research questions have grown more sophisticated. These methods provide limited, often biased information that fails to capture the structural, mechanical, and phenotypic complexity of microbial communities. Advanced techniques including atomic force microscopy, flow cytometry, and correlative imaging approaches offer powerful alternatives that enable researchers to connect nanoscale mechanical properties with three-dimensional biofilm architecture and population heterogeneity. For researchers investigating biofilm-related infections, antimicrobial resistance, and microbial ecology, adopting these advanced methodologies is essential for generating meaningful, clinically relevant data. The integration of these approaches, particularly the correlation of AFM nanomechanics with confocal imaging, represents the future of biofilm research, enabling unprecedented insights into the structure-function relationships that govern biofilm resilience and pathogenicity.
Superior Resolution and In-Situ Capability vs. Scanning Electron Microscopy (SEM)
The study of microbial biofilms represents a significant frontier in biomedical research and therapeutic development. Biofilms are structured communities of microorganisms encased in a self-produced extracellular polymeric substance (EPS) matrix, contributing up to 80% of persistent human infections [72]. Their resistance to conventional antibiotics can be up to 1,000-fold greater than their planktonic counterparts, primarily due to matrix protection, metabolic dormancy, and enhanced efflux-pump expression [72]. Understanding the intricate architecture and mechanical properties of biofilms is therefore crucial for developing effective therapeutic interventions. This pursuit has driven the adoption of advanced microscopy techniques, particularly Atomic Force Microscopy (AFM) and Scanning Electron Microscopy (SEM), each offering distinct capabilities for nanoscale characterization. Within the context of correlating AFM nanomechanics with confocal biofilm imaging research, this guide provides an objective comparison of these technologies, focusing specifically on their resolution capabilities and performance in biologically relevant environments.
Technical Comparison: AFM vs. SEM
Atomic Force Microscopy and Scanning Electron Microscopy represent fundamentally different approaches to high-resolution imaging. AFM, developed in the 1980s, operates by scanning a sharp tip on a cantilever across a surface and measuring the interaction forces, functioning much like a nanoscale phonograph [73]. In contrast, SEM, practiced since the 1960s, uses a focused beam of electrons to probe the sample surface, with detected electrons and X-rays used to construct an image [73]. The following comparison delineates their core technical characteristics.
Table 1: Fundamental Characteristics of AFM and SEM
| Characteristic | Atomic Force Microscopy (AFM) | Scanning Electron Microscopy (SEM) |
|---|---|---|
| Physical Basis | Physical interaction between a sharp probe and the sample surface [74] | Emission of electrons from the sample after being struck by a focused electron beam [74] |
| Resolution | ~1 nm (XY), <0.1 nm (Z) [74]; Can achieve atomic-level resolution [75] | ~1 nm (XY) [74] [75] |
| Environment | Vacuum, air, gas, or liquid [74] [73] | High vacuum (typically) [74] [73] |
| Information Dimension | Three-dimensional topography [74] | Two-dimensional projection with pseudo-3D capability [75] [73] |
| Material Sensitivity | Material-independent; measures mechanical properties [74] | Somewhat increases with atomic number; provides elemental composition data [74] [73] |
Table 2: Comparative Analysis of Performance and Applications
| Aspect | Atomic Force Microscopy (AFM) | Scanning Electron Microscopy (SEM) |
|---|---|---|
| Key Strengths | Superior Z-resolution; nanomechanical mapping; operates in physiological conditions [74] [76] | Excellent for large depth of field and rough surfaces; rapid imaging; elemental analysis [75] [73] |
| Sample Preparation | Minimal preparation; can image native hydrated samples [75] [73] | Often requires conductive coating, fixation, and dehydration, which can alter native structure [75] [77] |
| Throughput | Slower scanning speed; smaller scan areas [75] | Higher throughput for imaging larger areas [75] |
| Ideal for Biofilm Research | Real-time visualization of biofilm formation in liquid; quantitative measurement of elasticity and adhesion [78] [76] | High-resolution visualization of fixed, dehydrated biofilm surface architecture [58] [77] |
Experimental Data and Protocols
Documenting Superior Resolution and In-Situ Capability
Experimental data consistently validates AFM's superior vertical resolution and unique capability for in-situ imaging. A direct comparison study on synthetic nanoparticles (silica, gold, and polystyrene) demonstrated that while all three techniques (AFM, SEM, TEM) could characterize the samples, AFM provided high-contrast, three-dimensional topography independent of the nanoparticle material [74]. Critically, for soft biological samples like biofilms, AFM's ability to operate in liquid environments enables researchers to observe dynamic processes such as initial bacterial adhesion and subsequent biofilm development under near-physiological conditions, a feat not possible with conventional SEM without significant sample alteration [73] [78].
The protocol for in-situ AFM imaging of biofilms involves growing a biofilm on a suitable substrate (e.g., glass, silicone). The substrate is then mounted in the AFM liquid cell, submerged in an appropriate buffer or growth medium. Nanomechanical mapping is typically performed using a force-volume mode or a dynamic (tapping) mode to minimize sample disturbance. The sharp AFM probe (with a spring constant of ~0.1-1 N/m for soft samples) scans the surface, and the force-distance curves collected at each pixel are analyzed to generate simultaneous topographical and mechanical property maps (e.g., Young's modulus) [76].
In contrast, a standard protocol for SEM analysis of biofilms, as used in a recent study on Pseudomonas aeruginosa biofilms in tubular structures, requires extensive sample preparation [58]. After treatment, biofilms are fixed with 2.5% glutaraldehyde, dehydrated through a graded ethanol series (e.g., 30%, 50%, 80%, 100%), and then dried [58]. To make the non-conductive sample conductive for imaging, the dried biofilm is coated with a thin layer of gold/palladium using a sputter coater [74] [58]. While this process produces high-resolution surface images, the fixation, dehydration, and coating steps can introduce artifacts and do not preserve the native hydrated state of the EPS matrix.
Case Study: Biofilm Analysis in Tubular Structures
A 2025 study investigating shockwave treatment for biofilm disruption provides a clear example of SEM's application and its context within a broader analytical workflow [58]. The research aimed to assess the effect of shockwaves combined with antibiotics on Pseudomonas aeruginosa biofilms formed on the inner surface of silicone tubes.
Experimental Workflow:
- Biofilm Formation: P. aeruginosa was cultured and circulated through silicone tubes for 72 hours under dynamic conditions to simulate a realistic infection environment [58].
- Treatment: Established biofilms were treated with a shockwave system (120 pulses at 2 Hz), followed by exposure to ciprofloxacin [58].
- Multi-Modal Analysis: Bacterial viability was assessed via Colony-Forming Unit (CFU) counts and Confocal Laser Scanning Microscopy (CLSM) with live/dead staining. Biofilm detachment and structural integrity were evaluated using Crystal Violet (CV) staining and SEM [58].
Results and Correlation: The SEM analysis provided direct visual evidence of biofilm detachment, showing a removal of up to 97.5% of the surface area in the combined treatment group compared to the control [58]. This structural data correlated with a 40% decrease in bacterial viability (CFU counts) and a significant reduction in biofilm biomass (CV staining) [58]. This case illustrates how SEM serves as a powerful tool for qualifying structural changes in biofilms, with its findings validated by complementary techniques that assess viability and total biomass.
The Scientist's Toolkit: Key Reagents and Materials
Successful biofilm imaging and analysis rely on a suite of specialized reagents and materials. The following table details essential items for the experiments cited in this guide.
Table 3: Essential Research Reagents for Biofilm Imaging Experiments
| Reagent / Material | Function / Application | Example in Context |
|---|---|---|
| Silicone Tubes | Provides a tubular substrate for in-vitro biofilm formation under dynamic flow conditions, mimicking medical devices like catheters [58]. | Used as the substrate for growing P. aeruginosa biofilms in the shockwave treatment study [58]. |
| Glutaraldehyde (e.g., 2.5%) | A fixing agent that cross-links and preserves the structural proteins of biofilms, preparing them for electron microscopy [58] [77]. | Used to fix wound-edge biopsies and tubular biofilms prior to SEM processing to maintain structure [58] [77]. |
| Gold/Palladium Coating | A conductive metal layer applied to non-conductive biological samples via sputter coating to prevent charging and improve signal during SEM imaging [74] [58]. | Essential for obtaining high-quality SEM images of non-conductive polystyrene and silica nanoparticles, and fixed biofilms [74] [58]. |
| Ciprofloxacin | A fluoroquinolone antibiotic with strong activity against Pseudomonas aeruginosa and biofilm-associated strains; used to test antimicrobial efficacy [58]. | Applied at 4 µg/ml for 6 hours post-shockwave treatment to assess combined therapy efficacy [58]. |
| SYTO9/PI Staining Kit | A dual-fluorescence stain for differentiating live (green, SYTO9) and dead (red, Propidium Iodide) bacterial cells in a population, used with CLSM [58]. | Employed to visualize and quantify bacterial viability after combined shockwave and antibiotic treatment [58]. |
| Crystal Violet (CV) Stain | A colorimetric dye that binds to cells and extracellular polymeric substances, providing a quantitative measure of total biofilm biomass [58] [72]. | Used to stain treated biofilms in tubes, showing a significant reduction in biomass (OD600 of 0.14) [58]. |
| Ethanol Series | A graded sequence of ethanol concentrations (e.g., 30%, 50%, 80%, 100%) used to dehydrate biological samples after fixation for SEM analysis [58] [77]. | Critical step in preparing fixed biofilm samples for drying and SEM imaging [58] [77]. |
Integrated Workflow for Correlative AFM Nanomechanics and Confocal Biofilm Imaging
The true power of modern biofilm research lies in integrating multiple techniques to gain a comprehensive understanding. Correlating AFM nanomechanics with confocal imaging provides simultaneous data on the 3D structure, chemical composition, and mechanical properties of biofilms. AFM can determine the Young's modulus of a biofilm, providing quantifiable metrics on its stiffness or elasticity, which are critical for understanding its mechanical stability and response to treatment [76]. Concurrently, confocal microscopy can identify different bacterial species or visualize metabolic activity within the same region of interest using fluorescent tags.
This integrated approach allows researchers to link specific structural features or microbial communities identified by confocal microscopy with their local mechanical properties measured by AFM, offering unprecedented insights into biofilm heterogeneity and resilience mechanisms.
Both Atomic Force Microscopy and Scanning Electron Microscopy are indispensable tools in the arsenal of biofilm researchers. The choice between them is not a matter of which is universally superior, but which is most appropriate for the specific research question at hand. SEM excels in providing high-resolution, top-down visualizations of biofilm surface architecture with relatively high throughput, making it ideal for qualitative structural assessment and detachment studies. AFM, with its superior Z-resolution and unique ability to operate in liquid environments, provides unparalleled quantitative, three-dimensional topographical and nanomechanical data under near-physiological conditions. For research aimed at correlating structure with function—such as understanding how the mechanical properties of a biofilm contribute to its antibiotic resistance—the in-situ capability and nanomechanical mapping of AFM, especially when integrated with confocal microscopy, offers a powerful and often superior correlative approach.
Biofilms represent a significant challenge in medical, industrial, and environmental contexts due to their complex structure and remarkable resistance to antimicrobial treatments. This resilience stems from their heterogeneous architecture and adaptive capabilities, which vary significantly across different microbial species and environmental conditions. Understanding the spatial organization and mechanical properties of biofilms is crucial for developing effective control strategies. This case study objectively compares two advanced atomic force microscopy (AFM) approaches—large-area automated AFM and cell-level force-distance spectroscopy—for investigating biofilm heterogeneity and its correlation with substrate stiffness. By integrating nanomechanical data with structural insights, we demonstrate how these complementary AFM methodologies provide unique perspectives on biofilm cohesiveness and bacterial response to biomechanical cues, offering valuable insights for researchers and drug development professionals working at the intersection of AFM nanomechanics and confocal biofilm imaging.
Experimental Methodologies
Large-Area Automated AFM for Structural Analysis
Principle and Workflow: Traditional AFM imaging is limited by a narrow field of view (typically <100 μm), restricting researchers' ability to connect nanoscale features with larger biofilm architectures [7]. The large-area automated AFM approach overcomes this limitation by integrating automated scanning procedures with machine learning-assisted image analysis to capture high-resolution images across millimeter-scale areas [7] [79] [80].
Table 1: Key Protocol Steps for Large-Area Automated AFM
| Step | Procedure | Parameters | Purpose |
|---|---|---|---|
| Sample Preparation | Inoculate Pantoea sp. YR343 on PFOTS-treated glass coverslips | Incubation: ~30 min to 8h; Gentle rinsing before drying [7] | Ensure proper surface attachment while preserving native structures |
| Automated Scanning | Program AFM to capture multiple adjacent high-resolution images | Minimal overlap between scans; Millimeter-scale coverage [7] | Maximize acquisition speed while ensuring comprehensive area coverage |
| Image Stitching | Apply machine learning algorithms to merge individual scans | Limited feature matching between images [7] | Create seamless, high-resolution composite images |
| Data Analysis | Implement ML-based segmentation for parameter extraction | Cell detection, classification, and morphological analysis [7] [80] | Automate extraction of cell count, confluency, shape, and orientation |
Technical Innovations: This methodology leverages machine learning for multiple critical functions: optimizing scanning site selection to reduce human intervention, refining tip-sample interactions for improved image quality, and enabling automated probe conditioning for consistent performance [7]. The integration of sparse scanning approaches significantly reduces data acquisition time while maintaining resolution [7].
Cell-Level Force-Distance Spectroscopy for Nanomechanical Characterization
Principle and Workflow: Force-distance spectroscopy (FDS) based on AFM enables quantitative measurement of cell-surface adhesion forces and nanomechanical properties under physiological conditions [81] [5]. This approach quantifies how bacteria respond to variations in substrate stiffness, revealing species-specific adhesion mechanisms.
Table 2: Key Protocol Steps for Force-Distance Spectroscopy
| Step | Procedure | Parameters | Purpose |
|---|---|---|---|
| Substrate Preparation | Create LMP agarose pads with varying concentrations | Stiffness range: 20-120 kPa [81] | Modulate substrate stiffness in physiologically relevant range |
| Force Mapping | Perform array of force-distance curves across cell surfaces | Multiple measurements per cell; Controlled approach-retract cycles [81] [5] | Quantify adhesion forces at cell-substrate interface |
| Data Analysis | Apply mechanical models to force curves | Hertz model for elasticity; Adhesion force quantification [5] | Extract nanomechanical parameters including adhesion forces and Young's modulus |
| Control Experiments | Use polystyrene microparticles as reference | Comparable dimensions to bacterial cells [81] | Distinguish biological adhesion from passive interactions |
Technical Considerations: The Chen, Tu, and Cappella models, developed from the foundational Hertz model, are particularly suitable for thin biological samples on hard substrates [5]. For adhesion quantification, the analysis focuses on retract force-distance curves, where bond rupture events between the cell and surface provide direct measurement of adhesion forces [81] [5].
Comparative Data Analysis
Structural Heterogeneity Revealed by Large-Area AFM
The large-area automated AFM approach provided unprecedented insights into spatial organization during early biofilm formation. Analysis of Pantoea sp. YR343 biofilms revealed a distinctive honeycomb pattern of cellular organization, with preferential orientation of surface-attached cells [7] [80]. This methodology enabled automated analysis of over 19,000 individual cells across extensive surface areas, revealing several key structural features [79].
Table 3: Quantitative Structural Parameters from Large-Area AFM
| Parameter | Measurement | Significance |
|---|---|---|
| Cellular Dimensions | ~2 μm length, ~1 μm diameter [7] | Corresponds to surface area of ~2 μm², aligning with previous findings |
| Flagellar Structures | 20-50 nm height; Tens of micrometers extension [7] | Suggests role in biofilm assembly beyond initial attachment |
| Spatial Organization | Honeycomb-like pattern with characteristic gaps [7] [79] | Indicates coordinated cellular positioning during early development |
| Bacterial Density on Modified Surfaces | Significant reduction on silicon substrates [7] [80] | Highlights potential for surface modifications to control biofilm formation |
The high-resolution capability of this approach allowed clear visualization of flagellar structures bridging gaps between cells during early attachment phases [7]. These intricate appendages, confirmed through comparison with flagella-deficient control strains, appear to play a crucial role in intercellular coordination and biofilm maturation [7].
Nanomechanical Response to Substrate Stiffness
Force-distance spectroscopy experiments demonstrated a remarkable species-specific response to substrate stiffness variations. The research quantified how bacterial adhesion forces change with increasing substrate stiffness, revealing distinct adaptive strategies between bacterial species [81].
Table 4: Nanomechanical Adhesion Parameters from Force-Distance Spectroscopy
| Parameter | Chromatium okenii | Escherichia coli | Polystyrene Microparticles |
|---|---|---|---|
| Adhesion Range | 0.21±0.10 nN to 2.42±1.16 nN [81] | 0.29±0.17 nN to 0.39±0.20 nN [81] | No perceptible change [81] |
| Stiffness Dependence | Strong (over 10-fold increase) [81] | Weak | None |
| Biological Significance | Active modulation of adhesion as functional trait [81] | Limited stiffness sensitivity | Control for passive interactions |
Beyond interspecies variation, the study revealed phenotypic diversification within bacterial populations across growth stages. For Chromatium okenii, researchers observed the emergent co-existence of weakly and strongly adherent subpopulations over time, demonstrating how adhesion phenotypes diversify throughout bacterial growth cycles [81].
Integrated Analysis and Discussion
Correlating Structural and Nanomechanical Data
The combination of large-area structural mapping and nanomechanical characterization provides a comprehensive understanding of biofilm heterogeneity. The structural data obtained through large-area AFM explains how cellular patterns and flagellar networks contribute to macroscopic biofilm properties, while force spectroscopy reveals the fundamental mechanical interactions at the cellular level that enable such organization.
Diagram: Relationship between substrate stiffness and biofilm organization. Substrate properties modulate bacterial response, which determines adhesion forces that guide biofilm structure, creating potential feedback mechanisms.
The honeycomb pattern observed through large-area AFM [7] [79] likely represents an optimized architectural arrangement that maximizes mechanical stability while facilitating nutrient transport. This organization may be enabled by the stiffness-dependent adhesion mechanisms quantified through force spectroscopy [81], where bacteria actively modulate their attachment in response to biomechanical cues.
Implications for Biofilm Control Strategies
Understanding the interplay between structural heterogeneity and nanomechanical properties opens new avenues for biofilm control. The significant reduction in bacterial density observed on modified silicon substrates [7] [80] suggests that surface engineering approaches can effectively disrupt initial attachment. Similarly, the species-specific response to substrate stiffness [81] indicates that tailored surface modifications could selectively discourage colonization by particular pathogens while preserving beneficial microbiota.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 5: Key Research Reagent Solutions for AFM Biofilm Studies
| Item | Function | Application Notes |
|---|---|---|
| PFOTS-treated Glass Coverslips | Hydrophobic surface for bacterial attachment studies [7] | Enables observation of early attachment dynamics in Pantoea sp. YR343 |
| Low-Melting-Point (LMP) Agarose Pads | Tunable stiffness substrates for mechanical studies [81] | Allows stiffness modulation between 20-120 kPa to match physiological environments |
| Pantoea sp. YR343 | Model gram-negative bacterium for biofilm studies [7] | Rod-shaped, motile bacterium with peritrichous flagella; forms characteristic honeycomb patterns |
| Chromatium okenii | Phototrophic bacterium for adhesion studies [81] | Exhibits strong stiffness-dependent adhesion, increasing over 10-fold across stiffness range |
| Flagella-Deficient Control Strains | Reference for structural identification [7] | Confirms flagellar identity through absence of appendages in AFM imaging |
| Polystyrene Microparticles | Control for non-biological adhesion [81] | Demonstrates biological specificity of stiffness-dependent adhesion phenomena |
Methodological Integration Pathways
The complementary nature of large-area structural analysis and nanomechanical characterization suggests powerful opportunities for methodological integration in future biofilm research. Combined experimental approaches could simultaneously map structural heterogeneity and mechanical properties across multiple spatial scales.
Diagram: AFM methodological integration. Combining large-area AFM structural data with force spectroscopy mechanical measurements enables comprehensive biofilm insights through integrated data analysis.
This comparative analysis demonstrates that both large-area automated AFM and force-distance spectroscopy provide unique yet complementary insights into biofilm heterogeneity. The large-area approach excels at revealing structural patterns and spatial organization across clinically relevant scales, while force spectroscopy quantifies nanomechanical interactions at the cellular level. Together, these methodologies enable researchers to connect microscale structural features with nanoscale mechanical properties, offering a comprehensive framework for understanding biofilm cohesiveness in relation to substrate stiffness. For drug development professionals, these insights create opportunities for novel anti-biofilm strategies that target the mechanical aspects of biofilm formation and persistence, potentially overcoming limitations of conventional antimicrobial approaches. The continued integration of AFM nanomechanics with complementary imaging techniques like confocal microscopy will further advance our ability to correlate structural heterogeneity with functional outcomes in biofilm-associated infections and resistance mechanisms.
Biofilms are structured microbial communities encased in a self-produced extracellular polymeric substance (EPS) matrix, which represents a predominant mode of bacterial growth in both natural and clinical contexts [56]. This complex architecture confers remarkable resistance to antimicrobial agents and host immune responses, making biofilm-associated infections particularly challenging to treat [82]. The EPS matrix, composed of polysaccharides, proteins, nucleic acids, and lipids, creates a formidable physical and functional barrier that restricts antibiotic penetration, facilitates adaptive metabolic responses, and promotes persistent infection [83]. This protective environment increases bacterial resistance to antimicrobial treatments by up to 1000-fold compared to planktonic cells, contributing significantly to the global antimicrobial resistance (AMR) crisis [84] [85].
Quantifying the efficacy of anti-biofilm agents requires sophisticated methodologies that assess both their ability to penetrate this protective matrix and their mechanistic actions against specific biofilm components and pathways [82]. The therapeutic challenge is multifaceted: agents must first traverse the electrostatic, hydrophobic, and enzymatic barriers presented by the biofilm matrix, then effectively target bacterial cells often residing in heterogeneous metabolic states, and finally disrupt the structural integrity and signaling networks that maintain the biofilm community [56]. Understanding these complex interactions demands integrated analytical approaches, including advanced imaging techniques like Atomic Force Microscopy (AFM) and confocal laser scanning microscopy, which provide complementary data on structural modifications and functional outcomes at multiple scales [7].
Key Mechanisms of Biofilm Resistance to Antimicrobial Agents
The resilience of biofilms to conventional antibiotics stems from multiple interconnected resistance mechanisms that operate at physical, physiological, and genetic levels. The extracellular polymeric substance (EPS) matrix acts as a dual barrier, both physically impeding antibiotic penetration and chemically binding or degrading antimicrobial molecules through its constituent enzymes [85] [56]. This matrix creates a heterogeneous microenvironment with pronounced gradients of nutrients, oxygen, and metabolic waste products, leading to stratified bacterial subpopulations with varying metabolic activities [82]. Cells in nutrient-depleted zones enter slow-growing or dormant states, becoming intrinsically less susceptible to antibiotics that target active cellular processes [82].
Beyond these passive mechanisms, biofilms employ active defense strategies. Quorum sensing (QS) systems enable coordinated gene expression in a cell-density-dependent manner, regulating virulence factor production, matrix synthesis, and adaptive responses to environmental stresses [82] [83]. Biofilms also facilitate efficient horizontal gene transfer, accelerating the dissemination of antibiotic resistance genes among community members [85]. The oxidative stress response within biofilms provides additional protection against antimicrobial agents that generate reactive oxygen species, while the presence of persister cells - a small subpopulation of metabolically dormant bacteria - ensures community survival following antibiotic exposure, leading to recurrent infections [56]. This multi-layered resistance profile necessitates therapeutic approaches that simultaneously address multiple mechanisms rather than targeting single pathways.
Table 1: Primary Mechanisms of Biofilm-Associated Antimicrobial Resistance
| Resistance Mechanism | Functional Basis | Impact on Efficacy |
|---|---|---|
| Physical Barrier | EPS matrix limits antibiotic diffusion and penetration | Reduces intracellular drug concentration; requires higher doses |
| Metabolic Heterogeneity | Gradients create varied physiological states | Antibiotics targeting active processes fail against dormant cells |
| Enhanced Genetic Exchange | Close proximity facilitates plasmid transfer | Accelerates spread of resistance determinants |
| Quorum Sensing Regulation | Coordinate gene expression and stress responses | Enables community-wide adaptation to threats |
| Persister Cell Formation | Dormant subpopulation tolerant to antibiotics | Leads to treatment failure and recurrence |
Analytical Frameworks for Assessing Anti-biofilm Efficacy
Quantitative Assessment Metrics and Parameters
Standardized quantification of anti-biofilm activity requires multiple complementary metrics that capture different aspects of efficacy. The Minimum Biofilm Inhibitory Concentration (MBIC) represents the lowest concentration that prevents biofilm formation, while the Minimum Biofilm Eradication Concentration (MBEC) indicates the concentration required to eliminate mature biofilms [82]. These parameters differ significantly from conventional Minimum Inhibitory Concentration (MIC) measurements against planktonic cells, with MBEC values typically orders of magnitude higher than MIC values for the same organism-agent combination [82]. Additional quantitative measures include biofilm inhibition percentage, which calculates the reduction in biofilm biomass compared to untreated controls, and dispersal efficiency, which quantifies the release of previously attached cells [86].
Advanced analytical techniques provide deeper insights into anti-biofilm mechanisms. Metabolic activity assays using tetrazolium salts or ATP quantification distinguish between biocidal and biostatic effects, while crystal violet staining enables quantification of total biofilm biomass [87]. Confocal laser scanning microscopy (CLSM) with viability staining generates three-dimensional reconstructions of biofilm architecture and spatial patterns of live/dead cells following treatment [7]. When combined with computational analysis, these approaches can quantify changes in biofilm thickness, substratum coverage, and biovolume, providing comprehensive assessment of structural disruption [87]. Recently, large-area automated Atomic Force Microscopy (AFM) has enabled nanoscale resolution imaging of biofilm surface topography and mechanical properties across millimeter-scale areas, capturing structural heterogeneity previously obscured by conventional techniques [7].
Correlative AFM and Confocal Imaging Methodologies
The integration of AFM with confocal microscopy creates a powerful correlative framework that links nanomechanical properties with structural and functional information in biofilms. This combined approach enables researchers to map morphological changes at subcellular resolution while simultaneously monitoring bacterial viability and matrix composition in response to anti-biofilm treatments [7]. The exceptional resolution of AFM reveals ultrastructural features including individual cells, flagella, pili, and EPS matrix components, providing direct visualization of agent-induced modifications at the nanoscale [7] [69].
The experimental workflow begins with preparing biofilms on appropriate substrates optimized for both imaging techniques. Following treatment with anti-biofilm agents, samples are first imaged using CLSM with fluorescent markers for viability (e.g., SYTO 9/propidium iodide), specific matrix components, or reporter strains expressing fluorescent proteins [7]. The same regions of interest are then analyzed using AFM, which can operate in both air and liquid environments, with the latter preserving native hydration conditions [7]. Advanced implementations incorporate machine learning algorithms for automated image stitching, cell detection, and classification across large areas, enabling statistically robust analysis of structural heterogeneity and treatment effects [7]. This integrated methodology quantitatively links nanomechanical properties (stiffness, adhesion, viscoelasticity) with functional outcomes (viability, matrix integrity), providing unprecedented insight into structure-function relationships during anti-biofilm treatment.
Diagram 1: Correlative AFM-Confocal Biofilm Analysis Workflow. This integrated approach links nanomechanical properties with structural and functional information.
Mechanistic Classification of Anti-biofilm Agents
Targeted Therapeutic Approaches and Molecular Actions
Anti-biofilm agents employ diverse mechanistic strategies to disrupt the biofilm lifecycle, which can be broadly categorized into four primary modes of action. Matrix targeting agents directly degrade or disrupt the structural integrity of the EPS through enzymatic action (e.g., DNases, dispersin B, proteases) or chemical interactions that destabilize matrix components [88] [83]. Quorum sensing inhibitors interfere with bacterial cell-to-cell communication systems, preventing the coordinated gene expression necessary for biofilm maturation and virulence without exerting direct bactericidal pressure [87]. This anti-virulence approach includes molecules that degrade autoinducers, block receptor binding, or inhibit signal synthesis [87].
Nanoparticle-based systems leverage unique physicochemical properties to penetrate biofilm architecture and deliver concentrated antimicrobial payloads or generate localized reactive oxygen species [84] [85]. Metallic nanoparticles, particularly silver-based nanocomposites, exhibit broad-spectrum anti-biofilm activity through multiple mechanisms including membrane disruption, protein denaturation, and interference with enzymatic functions [86]. Natural product-derived compounds from terrestrial and marine sources represent a rich reservoir of structurally diverse anti-biofilm molecules, including phenolic compounds, flavonoids, essential oils, and antimicrobial peptides that often target multiple pathways simultaneously [83]. This polypharmacological approach is particularly valuable against resilient biofilm communities, as it reduces the likelihood of resistance development compared to single-target agents.
Table 2: Classification of Anti-biofilm Agents by Primary Mechanism of Action
| Mechanistic Class | Molecular Targets | Representative Agents |
|---|---|---|
| Matrix Disruptors | EPS components (eDNA, polysaccharides, proteins) | DNase I, proteases, dispersin B, chelating agents |
| Signaling Inhibitors | Quorum sensing systems (autoinducers, receptors) | Halogenated furanones, patulin, hamamelitannin analogs |
| Nanoparticle Systems | Bacterial membranes, multiple intracellular targets | Ag nanoparticles, liposomes, nanoemulsions, dendrimers |
| Natural Product Derivatives | Multiple targets including membranes and enzymes | Plant extracts, essential oils, antimicrobial peptides, biosurfactants |
| Cellular Function Disruptors | Metabolic pathways, cell division machinery | Conventional antibiotics (in combination), metallic oxides |
Signaling Pathways in Biofilm Formation and Inhibition
The transition from planktonic to biofilm lifestyle is orchestrated through complex regulatory networks that integrate environmental cues with intracellular signaling systems. The cyclic di-GMP (c-di-GMP) secondary messenger pathway serves as a central regulatory hub that controls the switch between motile and sessile behaviors in numerous bacterial species [82]. Elevated intracellular c-di-GMP levels promote biofilm formation through upregulation of adhesins and matrix production, while decreased concentrations favor dispersion and motility [82]. This signaling molecule binds to diverse receptors including transcription factors, enzymes, and riboswitches, making the enzymes responsible for its synthesis (diguanylate cyclases) and degradation (phosphodiesterases) attractive targets for anti-biofilm interventions.
The quorum sensing (QS) network represents another key signaling pathway that coordinates community behaviors in a cell-density-dependent manner [83]. Gram-negative bacteria typically employ acyl-homoserine lactone (AHL) signaling molecules, while Gram-positive species use autoinducing peptides, with both systems converging on regulation of collective behaviors including biofilm maturation, virulence factor production, and antibiotic tolerance [87]. Anti-biofilm strategies that target QS include AHL-lactonases and -acylases that degrade signals, competitive antagonists that block receptor binding, and antibodies that sequester autoinducers [87]. Additionally, two-component signal transduction systems enable bacteria to sense and respond to environmental stresses, often activating biofilm formation as a protective response; small molecule inhibitors of these histidine kinase-response regulator systems can prevent this adaptive resistance mechanism [82].
Diagram 2: Biofilm Signaling Pathways and Anti-biofilm Intervention Points. Key regulatory systems and their corresponding inhibitory strategies.
Quantitative Comparison of Anti-biofilm Agent Efficacy
Performance Metrics Across Agent Classes
Systematic comparison of anti-biofilm efficacy requires standardized assessment of multiple performance parameters across different agent classes. Nanoparticle-based formulations consistently demonstrate superior penetration capabilities due to their small size and tunable surface properties, with silver nanocomposites showing particularly strong performance against clinically relevant biofilms [86]. In recent studies, Ag/AgCl nanocomposites synthesized using Prunus mahaleb fruit pericarp extracts exhibited remarkable biofilm inhibition percentages up to 145.7%, significantly outperforming many conventional antibiotics [86]. However, cytotoxicity varies considerably within this class, with Ag/AgO and Ag/Ag₂O nanocomposites demonstrating substantially higher toxicity (LC₅₀ = 28 μg/ml in brine shrimp assays) compared to Ag/AgCl variants (LC₅₀ > 300 μg/ml) [86].
Natural product-derived agents offer favorable safety profiles while exhibiting moderate to strong anti-biofilm activity, often through multi-target mechanisms that reduce resistance development. Plant extracts, essential oils, and antimicrobial peptides typically achieve biofilm inhibition ranging from 50-80% at sub-MIC concentrations, with enhanced efficacy when used in combination with conventional antibiotics [83]. Quorum sensing inhibitors demonstrate potent activity against biofilm maturation and virulence without exerting direct bactericidal pressure, making them valuable components in combination therapies [87]. Computational approaches including molecular docking and molecular dynamics simulations have become invaluable for predicting compound-target interactions and optimizing QS inhibitor design prior to experimental validation [87].
Table 3: Quantitative Efficacy Comparison of Anti-biofilm Agent Classes
| Agent Category | Typical MBIC Range (μg/mL) | Penetration Efficiency | Cytotoxicity Profile | Resistance Potential |
|---|---|---|---|---|
| Silver Nanoparticles | 1-50 [86] | High (size-dependent) | Variable (LC₅₀: 28->300 μg/ml) [86] | Low (multiple targets) |
| Natural Products | 10-200 [83] | Moderate to High | Generally low | Low to Moderate |
| QS Inhibitors | 5-100 [87] | High (small molecules) | Typically low | Very low (non-biocidal) |
| Conventional Antibiotics | 10-1000+ [82] | Low (matrix restricted) | Variable (therapeutic index) | High (single targets) |
| Enzymatic Dispersers | 0.1-50 [88] | High (matrix degrading) | Generally low | Low |
Experimental Protocols for Efficacy Assessment
Standardized methodologies for evaluating anti-biofilm activity ensure reproducible and comparable results across studies. The microtiter plate crystal violet assay represents the most widely employed screening method for quantifying biofilm formation and inhibition [87] [86]. The protocol begins with inoculating bacterial suspensions in growth media with or without test compounds in 96-well plates, followed by incubation under appropriate conditions (typically 37°C for 24-48 hours) to allow biofilm development [86]. After incubation, planktonic cells are removed by gentle washing, and adherent biofilms are fixed with methanol or ethanol before staining with 0.1% crystal violet solution for 15-30 minutes [86]. Excess stain is removed by washing, and the bound dye is solubilized with ethanol or acetic acid before measuring optical density at 570 nm using a microplate reader [86]. Biofilm inhibition percentage is calculated using the formula: % inhibition = [(C - B) - (T - B)] / (C - B) × 100, where C is the OD₅₇₀ of positive control, B is OD₅₇₀ of negative control, and T is OD₅₇₀ of test compound [86].
For more sophisticated mechanistic studies, confocal microscopy coupled with live/dead staining provides spatial resolution of bactericidal activity within the biofilm architecture [7]. Biofilms are grown on appropriate surfaces (often glass coverslips or specialized imaging chambers), treated with test compounds, and then stained with fluorescent viability markers such as SYTO 9 and propidium iodide [7]. Image acquisition using confocal microscopy generates Z-stacks that can be reconstructed into three-dimensional representations of the biofilm, with subsequent computational analysis quantifying biovolume, thickness, substratum coverage, and viability ratios [7]. When combined with large-area automated AFM, this approach correlates structural and mechanical properties with biological activity, revealing nanoscale alterations in biofilm topography, stiffness, and adhesion following treatment [7]. This integrated protocol enables comprehensive assessment of both eradication efficacy and mechanistic actions, bridging the gap between bulk population measurements and single-cell analyses.
Research Reagent Solutions for Anti-biofilm Studies
Table 4: Essential Research Reagents and Materials for Anti-biofilm Investigations
| Reagent Category | Specific Examples | Research Applications |
|---|---|---|
| Biofilm Staining Reagents | Crystal violet, SYTO 9/propidium iodide (Live/Dead), Congo red | Biomass quantification, viability assessment, matrix visualization |
| Matrix Degrading Enzymes | DNase I, dispersin B, proteases (proteinase K, trypsin) | EPS disruption studies, penetration enhancement, mechanistic analysis |
| Nanoparticle Systems | Silver nanocomposites, liposomes, polymeric nanoparticles | Drug delivery evaluation, penetration mechanism studies, combination therapies |
| QS Signaling Modulators | AHL analogs, furanones, hamamelitannin derivatives | Signaling pathway analysis, anti-virulence screening, combination approaches |
| Imaging Substrates | PFOTS-treated glass, silicon wafers, flow cell systems | AFM and confocal microscopy, surface attachment studies, real-time monitoring |
| Computational Tools | Molecular docking software, MD simulation packages, image analysis algorithms | In silico screening, mechanism prediction, quantitative image analysis |
The quantitative assessment of anti-biofilm agent efficacy requires multidisciplinary approaches that integrate penetration metrics with mechanistic actions across multiple scales. The correlative framework combining AFM nanomechanics with confocal imaging represents a powerful methodology for linking structural and mechanical modifications with biological outcomes, providing unprecedented insights into anti-biofilm mechanisms [7]. As biofilm research continues to evolve, several emerging trends are shaping future directions: the development of smart nanoparticle systems with stimuli-responsive properties that enable targeted drug release in specific biofilm microenvironments [84] [85]; the increasing application of machine learning and artificial intelligence for automated image analysis, predictive modeling, and high-throughput data processing [7]; and the strategic implementation of combination therapies that simultaneously target multiple vulnerability points in the biofilm lifecycle [83].
Future advances in anti-biofilm therapeutics will depend on continued refinement of quantitative assessment methodologies, particularly those that bridge the gap between in vitro models and clinical reality. Standardization of testing protocols across laboratories will enhance comparability between studies, while development of more sophisticated in vivo imaging techniques will improve translational predictability [88]. The integration of computational approaches with experimental validation creates exciting opportunities for rational design of next-generation anti-biofilm agents with optimized penetration properties and mechanism-based efficacy [87]. As these methodologies mature, they will accelerate the development of effective therapeutic strategies against persistent biofilm-associated infections, ultimately addressing a critical unmet need in antimicrobial therapy.
Conclusion
The integration of AFM nanomechanics with confocal biofilm imaging represents a paradigm shift in biofilm research, moving beyond static structural analysis to a dynamic, functional understanding. This correlative approach uniquely reveals how the nanoscale mechanical properties of a biofilm, such as localized stiffness and cohesive strength, directly dictate its 3D architecture and formidable resistance. The key takeaway is that this multimodal framework provides unprecedented insights into structure-function relationships, enabling the rational design of targeted anti-biofilm strategies that disrupt mechanical integrity. Future directions will be shaped by emerging trends, including the increased use of AI for automated analysis of large, multimodal datasets, the development of real-time correlative imaging under fluidic conditions to observe dynamic processes, and the application of these insights to engineer smart nanomaterials and novel enzymatic therapies capable of precisely degrading the biofilm matrix. This paves the way for a new era of precision-guided biofilm management in both clinical and industrial settings.
References
- 1. Biofilms Exposed: Innovative Imaging and Therapeutic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12466655/]
- 2. Molecular imaging of bacterial biofilms—a systematic review [https://pmc.ncbi.nlm.nih.gov/articles/PMC11523921/]
- 3. Review: Advanced Atomic Force Microscopy Modes for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9775674/]
- 4. Atomic force microscopy-mediated mechanobiological ... [https://www.sciencedirect.com/science/article/pii/S0142961223003976]
- 5. Nanomechanical characterization of soft nanomaterial ... [https://www.sciencedirect.com/science/article/pii/S259000642500064X]
- 6. Pili and other surface proteins influence the structure ... [https://www.nature.com/articles/s41598-021-84030-1]
- 7. Analysis of biofilm assembly by large area automated AFM [https://www.nature.com/articles/s41522-025-00704-y]
- 8. Confocal microscopy of oral streptococcal biofilms grown in ... [https://www.nature.com/articles/s41526-024-00427-y]
- 9. Comparison of microscope techniques for the examination ... [https://www.sciencedirect.com/science/article/abs/pii/0167701295000852]
- 10. Artificial intelligence-empowered imaging technologies for ... [https://www.sciencedirect.com/science/article/abs/pii/S1385894725084311]
- 11. Advances in nanomechanical property mapping by atomic ... [https://pubs.rsc.org/en/content/articlehtml/2025/na/d5na00702j]
- 12. Nanomechanical characterization of soft nanomaterial ... [https://pubmed.ncbi.nlm.nih.gov/40018054/]
- 13. Atomic Force Microscopy (AFM) nanomechanical ... [https://www.sciencedirect.com/science/article/pii/S2405665025000125]
- 14. Abstract [https://arxiv.org/html/2509.15242v1]
- 15. Deciphering conformational dynamics in AFM data using ... [https://www.nature.com/articles/s42003-025-08365-5]
- 16. Flexible Fitting to Infer Atomistic-Precision Models of Large ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12490010/]
- 17. Confocal Laser Scanning Microscopy - an overview [https://www.sciencedirect.com/topics/neuroscience/confocal-laser-scanning-microscopy]
- 18. Quantitative evaluation of the site-dependent cell viability in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11934154/]
- 19. Confocal Laser Scanning Microscopy - an overview [https://www.sciencedirect.com/topics/chemistry/confocal-laser-scanning-microscopy]
- 20. Confocal laser scanning microscopy reveals species ... [https://www.frontiersin.org/journals/marine-science/articles/10.3389/fmars.2024.1483206/full]
- 21. Challenges in 3D Live Cell Imaging [https://www.mdpi.com/2304-6732/8/7/275]
- 22. Simultaneous co-localized super-resolution fluorescence ... [https://www.nature.com/articles/s41598-020-57885-z]
- 23. Confocal Laser Scanning Microscope Imaging of Custom ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10221725/]
- 24. Microscopy Methods for Biofilm Imaging: Focus on SEM ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7828176/]
- 25. AFM-Based Correlative Microscopy Illuminates Human ... [https://www.frontiersin.org/journals/cellular-and-infection-microbiology/articles/10.3389/fcimb.2021.655501/full]
- 26. Imaging of bacterial multicellular behaviour in biofilms ... [https://www.nature.com/articles/srep25889]
- 27. Confocal laser scanning microscopy as a tool to validate ... [https://www.sciencedirect.com/science/article/abs/pii/S1383586613006874]
- 28. A Multi-scale Biophysical Approach to Develop Structure ... [https://www.nature.com/articles/s41598-018-23798-1]
- 29. Monitoring of biofilms grown on differentially structured ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6999451/]
- 30. Methods to probe the formation of biofilms: applications in ... [https://pubs.rsc.org/en/content/articlehtml/2020/ay/c9ay02214g]
- 31. Multidimensional Atomic Force Microscopy: A Versatile Novel ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2976997/]
- 32. Soft matter analysis via atomic force microscopy (AFM) [https://www.sciencedirect.com/science/article/pii/S266652392300082X]
- 33. Biofilm Analysis by Confocal Microscopy-Basics and ... [https://pubmed.ncbi.nlm.nih.gov/40927881/]
- 34. Near Simultaneous Laser Scanning Confocal and Atomic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7680637/]
- 35. Advances in nanomechanical property mapping by atomic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12379807/]
- 36. Application of atomic force microscopy in cancer research [https://jnanobiotechnology.biomedcentral.com/articles/10.1186/s12951-018-0428-0]
- 37. A comprehensive study of AFM stiffness measurements on ... [https://www.nature.com/articles/s41598-024-75958-1]
- 38. Absolute Quantitation of Bacterial Biofilm Adhesion and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2711294/]
- 39. Atomic Force Microscopy Analysis of Polymer Stiffness [https://www.azonano.com/article.aspx?ArticleID=6890]
- 40. Quantification of bacterial adhesion forces using atomic ... [https://www.sciencedirect.com/science/article/abs/pii/S0167701299001372]
- 41. Biofilm Cohesiveness Measurement Using a Novel Atomic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC1892862/]
- 42. Biofilm viability checker: An open-source tool for automated ... [https://www.nature.com/articles/s41522-021-00214-7]
- 43. Key issues in biofilm reactors and AI-driven performance ... [https://www.sciencedirect.com/science/article/pii/S2666498424000942]
- 44. R‐based method for quantitative analysis of biofilm thickness ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9162930/]
- 45. New imaging approach transforms study of bacterial biofilms [https://www.ornl.gov/news/new-imaging-approach-transforms-study-bacterial-biofilms]
- 46. Biofilm analysis by confocal laser scanning microscopy [https://bio-protocol.org/exchange/minidetail?type=30&id=9668973]
- 47. afmbiomed 2025 - IBEC Events [https://events.ibecbarcelona.eu/afm-biomed-2025/]
- 48. What does 2025 have in store for AFM? - NuNano [https://www.nunano.com/blog/2025/1/26/what-does-2025-have-in-store-for-afm]
- 49. Atomic Force Microscopy‐Based Nanomechanical Signatures ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12590530/]
- 50. Nonlinear Harmonics: A Gateway to Enhanced Image ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11923995/]
- 51. Advances in multimodal imaging techniques in nanomedicine [https://pmc.ncbi.nlm.nih.gov/articles/PMC12308829/]
- 52. A data management infrastructure for the integration of ... [https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-022-04584-3]
- 53. A guide for nanomechanical characterization of soft matter ... [https://www.sciencedirect.com/science/article/pii/S2666166725002151]
- 54. Bone mineral tessellation: Atomic force microscopy of the ... [https://www.sciencedirect.com/science/article/pii/S1742706125003411]
- 55. Biofilm formation and analysis of EPS architecture ... [https://www.sciencedirect.com/science/article/abs/pii/S0963996925006118]
- 56. Biofilms: structure, resistance mechanism, emerging control ... [https://pubs.rsc.org/en/content/articlehtml/2025/pm/d5pm00094g]
- 57. applications of atomic force microscopy in disease [https://pmc.ncbi.nlm.nih.gov/articles/PMC12075069/]
- 58. Investigation of shockwave treatment for disruption ... [https://www.nature.com/articles/s41598-025-19955-y]
- 59. AFM-Based Correlative Microscopy Illuminates Human ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8138568/]
- 60. Confocal Microscopy: Principles and Modern Practices - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6961134/]
- 61. Correlative atomic force microscopy quantitative imaging ... [https://www.nature.com/articles/s41598-018-26433-1]
- 62. Near Simultaneous Laser Scanning Confocal and Atomic Force Microscopy (Conpokal) on Live Cells - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7680637/]
- 63. High-resolution imaging using a novel atomic force microscope and confocal laser scanning microscope hybrid instrument: essential sample preparation aspects - PubMed [https://pubmed.ncbi.nlm.nih.gov/18719934/]
- 64. Deep learning strategy for small dataset from atomic force ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10582561/]
- 65. Atomic Force Microscopy of Biofilms—Imaging, Interactions ... [https://www.intechopen.com/chapters/50739]
- 66. Quantitative and Qualitative Assessment Methods for Biofilm ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6133255/]
- 67. Crystal Violet Staining Alone Is Not Adequate to Assess ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8766793/]
- 68. Quantitative assessment of bacterial growth phase utilizing ... [https://www.sciencedirect.com/science/article/abs/pii/S0167701219307560]
- 69. AFM – Delivering Profound Insights in Microbiology [https://www.bruker.com/en/news-and-events/webinars/2025/afm-delivering-profound-insights-in-microbiology.html]
- 70. Toward an Improved In Vitro Model of Prosthetic Joint ... [https://www.sciencedirect.com/science/article/pii/S2590207525000735]
- 71. Comparison of Two Models of Biofilm Formation on ... [https://pubmed.ncbi.nlm.nih.gov/41265798/]
- 72. Biofilms Exposed: Innovative Imaging and Therapeutic ... [https://www.mdpi.com/2079-6382/14/9/865]
- 73. The Differences Between Atomic and Scanning... Force Microscopy [https://www.azom.com/article.aspx?ArticleID=11879]
- 74. , Comparing and TEM AFM SEM [https://www.afmworkshop.com/newsletter/comparing-afm-sem-and-tem]
- 75. Atomic Force Microscopy Vs SEM: Resolution, Cost, ... [https://eureka.patsnap.com/report-atomic-force-microscopy-vs-sem-resolution-cost-efficiency]
- 76. Nanomechanical Mapping with AFM ; Considering Cellular Structures [https://www.azonano.com/article.aspx?ArticleID=6043]
- 77. SEMTWIST Quantification of Biofilm Infection in Human ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12359142/]
- 78. Advances in imaging techniques for real-time microbial ... [https://www.sciencedirect.com/science/article/abs/pii/S0165993625000950]
- 79. DOE Scientists Develop Novel AFM Method to Transform ... [https://www.labcompare.com/617-News/620684-DOE-Scientists-Develop-Novel-AFM-Method-to-Transform-Biofilm-Research/?catid=61]
- 80. Analysis of biofilm assembly by large area automated AFM [https://pubmed.ncbi.nlm.nih.gov/40341406/]
- 81. The Dual Role of Substrate Stiffness and Bacterial Growth ... [https://pubmed.ncbi.nlm.nih.gov/40142529/]
- 82. Strategies for combating bacterial biofilms: A focus on anti ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5955472/]
- 83. Recent Progress in Terrestrial Biota-Derived Anti-Biofilm ... [https://www.mdpi.com/2673-8007/4/3/94]
- 84. Nanoparticles targeting biofilms: A new era in combating ... [https://www.sciencedirect.com/science/article/pii/S2590097825000370]
- 85. Nanomaterial-enabled anti-biofilm strategies - RSC Publishing [https://pubs.rsc.org/en/content/articlehtml/2025/nr/d4nr04774e]
- 86. Comparison of anti-biofilm and cytotoxic activity of Ag/AgO, ... [https://www.nature.com/articles/s41598-025-11756-7]
- 87. Integrative Experimental and Computational Dataset for ... [https://www.sciencedirect.com/science/article/pii/S2352340925007474]
- 88. Antibiofilm Agents for the Treatment and Prevention of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11420799/]